Medical School Expert

Medicine Essay Prizes (7 Competitions For Year 12 and 13’s)
Every article is fact-checked by a medical professional. However, inaccuracies may still persist.
Having a medical essay prize on your CV looks absolutely fantastic when it comes time for you to apply to medical school.
In such an overcrowded marketplace, anything that sets you apart from the crowd in a positive manner is sure to drastically increase your chances of getting an offer.
Although when I was applying to medical school I hadn’t managed to win an essay competition (despite my best efforts!) hopefully you’ll have more luck than me!
To save you some research time, I’ve compiled a list of 7 medicine essay competitions that you can enter this year.
Competitions that if you win will skyrocket your chances of application success.
INCLUDED IN THIS GUIDE:
The Libra Essay Prize
| Open to | Years 12 & 13 |
| Word count | 1,500 – 2,000 words |
| Prize | £50 |
The Libra essay prize is an annual essay prize for all students in years 12 and 13 looking to prepare for university.
Libra Education themselves describe it as:
“An excellent way for 6th-form students to demonstrate that they have the makings of a scholar, the Libra Essay Prize offers a chance to prepare for the academic rigour required by university assignments and provides a great accomplishment to discuss on a personal statement or at an interview.”
Students are free to choose any subject from a list of categories (one of which being science) and then have to write an essay with a title containing a chosen word.
The small pool of words you can choose from change each year but are all generally quite abstract so you can connect and use them in creative ways.
The essay has to be between 1,500 – 2,000 words, with Harvard style referencing which isn’t included in the word count.
Libra accept entries from all over the world, but the essays must be written in English.
First prize wins £50, second prize £30, and third £20, all paid out in book vouchers. There’s also Commended and Highly Commended entries for each category.
Minds Underground Essay Competition
| Open to | Year 12 and younger |
| Word count | 1,000 – 1,500 words |
| Prize | £30 voucher |
Minds Underground is an online learning platform, designed to support and enhance the learning and problem-solving of determined young students.
Every year they run a medicine essay competition, primarily aimed at year 12’s (although they do say younger or older students are also welcome to apply).
To enter, you have a choice of three questions, to which you need to write a 1,000 – 1,500 word answer.
“Should all healthcare be free? Discuss.” “What goes wrong for cancers to develop?” “Tell us about a key development/invention that you think has been most influential to medicine.” – Past Minds Underground medicine essay questions
If you’re feeling ambitious, students are permitted to enter an essay for more than one subject- so you could have a crack at the psychology or science one too!
Helpfully, under each question Minds also give you a few pointers to get your creative juices flowing.
Newnham Essay Prizes
| Open to | Female students in year 12 |
| Word count | 1,500 – 2,500 words |
| Prize | £400 |
Newnham College of the University of Cambridge runs a medicine essay competition with a twist:
Only female students are allowed to enter.
Again, students have a choice of three differing questions.
For example, the questions in the 2021-22 competition were:
- How realistic is it to develop a small molecule therapy for Covid-19? Could such a therapy be rolled out in a timeframe that it could have an impact on the current pandemic?
- Sleep deprivation in clinical health settings. Does it matter?
- Looking to the future. Will stem cell therapies be outpaced by machine-brain interfaces for the treatment of retinal disease?
Newnham do give you a bit more of a range when it comes to the word count, accepting anything from 1,500 to 2,500 words.
There’s a generous £400 prize for first place, £200 for second and £100 for third.
Unfortunately for you as an individual though, prize money is split 50:50 between the essay prize winner and the funding of resources for their school…
John Locke Institute Essay Competition
| Open to | Candidates must be 18 years old or younger |
| Word count | Less than 2,000 words |
| Prize | A scholarship worth $2,000 |
“The John Locke Institute encourages young people to cultivate the characteristics that turn good students into great writers: independent thought, depth of knowledge, clear reasoning, critical analysis and persuasive style. “
The John Locke Institute arguably gives away the most generous prize out of any competition on this list.
You get a scholarship worth $2,000 towards the cost of attending any John Locke Institute program, as well as an invitation to their prize-giving ceremony in Oxford.
The essay questions for each subject are published in January, with the deadline for submission generally being in late June.
As well as the opportunity to secure the prize for medicine, the candidate who submits the best essay overall will be awarded an honorary John Locke Institute Junior Fellowship- which comes with a $10,000 scholarship to attend one or more of their courses!
American Society Of Human Genetics Essay Contest
| Open to | Students in grades 9-12 |
| Word count | Less than 750 words |
| Prize | $1,000 for the student $1,000 genetics materials grant |
Although this next essay competition comes from America, it’s open to students worldwide.
The American Society Of Human Genetics supports national DNA day through its annual DNA day essay contest: commemorating the completion of the Human Genome Project in April 2003 and the discovery of the double helix of DNA in 1953.
The contest is open to students in grades 9-12 worldwide and asks students to “examine, question, and reflect on important concepts in genetics.”
With a limit of only 750 words, not including reference lists, this is a short but sweet chance to bag yourself a considerable cash prize for your efforts.
In addition to the personal prize money, the ASHG will also provide you with a $1,000 grant towards genetics research or teaching materials.
Although it is a worldwide contest, so undoubtedly will have plenty of entries, there are also 10 honorable mentions up for grabs (in addition to 1st, 2nd and 3rd place) that all come with a $100 prize too.
Immerse Education Essay Competition
| Open to | People aged 12-18 |
| Word count | Less than 500 words (+/- 10%) |
| Prize | 100% scholarship to Immerse summer school |
Immerse Education run summer programs for over 20 different subjects in Oxford, Cambridge, London and Sydney.
The reason why students get so much value from these courses is because they’re immersed in centers of academic excellence whilst learning from experts in their chosen field.
The good news for you is that their essay competition gives you the chance to attend one of their summer school programs for free.
10 winners receive a 100% scholarship and runners up are awarded partial scholarships of up to 50% to study their chosen subject.
According to Immerse, around 7% of entrants receive scholarship funding to attend a program- which is pretty good odds if you ask me!
“There is no downside to entering the competition. If you win, it is awesome. If you don’t win, you gained an experience. Entering the competition and working as hard as I did for it was one of the most gratifying experiences.” – Pedro L (100% scholarship winner)
You can find the full list of essay questions, in addition to top tips for writing academic essays (as well as the terms and conditions for the competition), simply by signing up via Immerse’s website.
The RCSU Science Challenge
| Open to | UK years 10-13 |
| Word count | Less than 1,000 words |
| Prize | Varies from year to year |
The Royal College of Science Union (RCSU) is a student union at Imperial College London and run an annual science challenge open to both home and international school students.
The focus of the challenge is communicating scientific concepts in a non-technical manner, so that people without a science background could still understand and enjoy the content.
The big twist with this essay competition is that you don’t actually have to enter an essay!
The idea is to produce a ‘short piece of science communication’ which can be an essay or can be a short video in answer to one of the four questions set by the judges.
Written entries must be less than 1,000 words, whilst video entries must be less than 3 minutes and 30 seconds long.
For this competition, it’s really all about short and snappy responses that will captivate the reader whilst answering the question in a precise but easy to understand manner.
We hope to inspire those who take part in the Science Challenge to explore, develop and use their scientific skills along with their passion for their corner of science to help others see what all the excitement is about.
Why You Should Enter Medicine Essay Competitions
I think it’s fair to say that competition to get into medical school in the UK is insanely high- and it’s only getting worse.
With such large numbers of incredibly qualified candidates, medical schools have to find some way of differentiating them.
One way to make it easy for a university to pick you is to stand out from the crowd by having a medical essay prize on your application.
An essay prize demonstrates your dedication to the subject, scientific knowledge and an ability to write expressively and persuasively- all ideal qualities when it comes to being a doctor.
You may surprise yourself.
Often, not as many people as you might think enter these competitions.

Simply by writing the essay, you’re also going to greatly increase your knowledge about that particular topic, which can still come in really handy at interview.
Even if you don’t win, just discussing the fact you entered still looks good in the eyes of an interviewer.
It shows that you’re willing to go above and beyond your school curriculum, to explore subjects you’re interested in and that you’re a highly motivated candidate.
How To Increase Your Chances Of Winning An Essay Prize
Although when I was applying to medical school I didn’t manage to win an essay prize, there are a couple of things I did that would have greatly increased my chances of doing so.
First and foremost, I think you’ve got to cast your net wide.
Don’t limit yourself to just one shot at the target: if you’ve got the time then I’d recommend trying to enter at least a couple of different competitions.
More entries will mean more chances for you to have your essay officially recognised.
Secondly, if you have the choice between entering a local or national competition, I’d always go with the local one.
Although a national prize would look slightly better on your CV, simply due to the number of entries, you’ll have a much higher chance of winning the more local competition.
By local I mean this could be a more regional charity, nearby hospital or university, or even your school.
Even better yet, you could always enter both!
Lastly, I think one of the best ways you can increase your odds of winning a prize is by entering a competition around a topic that you’re genuinely passionate about.
If you’ve no interest in genetics, then I wouldn’t enter the American Society Of Human Genetics’ contest!
Your interest in the subject will come through in your language, depth of knowledge and motivation to go above and beyond for your essay- all of which will put you in a much better position for winning.
Where You Can Find Further Essay Competitions
In addition to the essay prizes described above, there are tonnes of other opportunities available for you to distinguish yourself as a medicine applicant.
Loads of the Royal Colleges run an ever changing variety of prizes and competitions, usually to encourage interest in their specialty.
The opening dates and deadlines for these prizes are always changing so it’s worth keeping an eye out for the perfect essay question or new prize that’s just been announced.
Some of these organisations that run their own competitions include:
- The Royal Society of Medicine
- Royal College of Emergency Medicine
- British Orthopaedic Association
- British Society for Haematology
- Royal College of Pathologists
- Royal College of Psychiatrists
- British Association of Dermatologists
But there are many more out there. If you have a particular interest in one specialty or area of science then I’d definitely recommend doing a bit of digging to see if there’s a society or organisation related to that field that runs their own competitions!
Final Thoughts
There really aren’t many downsides to entering one of these competitions.
You get a shot at winning, gain a talking point at interview and develop your scientific knowledge (not to mention technical writing skills).
Although you might feel that some of the smaller prizes aren’t worth your time and effort to write the essay, the real value comes from the boost one of these prizes would give your medicine application.
What UCAT Score Is Needed For Medicine? (UK Specific)
Best Podcasts For Medicine Applicants (11 Must Listen Shows)
Convert your interviews into offers
Learn the best ways of turning your upcoming interviews into medicine offers here.
© 2024 Medical School Expert Ltd
Username or email *
Password *
Forgotten password?
[email protected]
+44 (0)20 8834 4579
School Competitions And Prizes For Aspiring Medics
Learn about all of the competitions and prizes you can enter at school to boost your Medicine application.
If you’re an aspiring medic at school, you can boost your Medicine application by entering competitions and prizes. Regardless of whether you win or not, you’ll be able to include the experience in your Personal Statement and talk about it at Med School interviews . Here are some Medicine competitions you can enter to be proactive and make your application stand out.
Imperial College London – Science in Medicine School Teams Prize
Imperial College London has three team competitions to choose from:
- The British Heart Foundation Cardiovascular Prize – Submissions should focus on a topic with interactions between the cardiovascular system and the nervous system.
- The Lung Prize – Submissions can focus on any aspect of the prevention or treatment of respiratory disease.
- The Scleroderma and Raynaud’s UK Prize – Submissions should focus on promoting the health and wellbeing of individuals with Scleroderma and/or Raynaud’s.
For each competition, the challenge is to design an ePoster. A team can have up to six members (they recommend assembling a team with varied interests) and schools can enter one team per prize.
The top ten shortlisted teams in each contest will be invited to present their ePosters at an online finals event. In each stream, first, second and third prizes of £3,000, £2,000 and £1,000 will be awarded to schools in order to support science-related activities.
Deadline – midnight on 30th June 2023.
University of Cambridge – Robinson College Essay Prize
The Robinson College Essay Prize is open to Year 12 students in the UK, providing an opportunity to develop and showcase independent study and writing skills. It also allows students to experience the type of work that they might be expected to do at Cambridge.
Entrants submit an essay (no more than 2,000 words) answering a question from various options. Last year, one of the possible titles was ‘Can science tell us how we should live?’. Up to three entries can be submitted per school, so you should discuss your application with your school before entering.
Five prizes are awarded, with each winner receiving book tokens to the value of £50. Winners will also be invited to Robinson College for a prize-giving ceremony.
The 2023 prize will open with more info in June.
Specialist Application Advice
Want expert advice to navigate the Medicine application process?
Medic Mentor – National Essay Competition
Medic Mentor’s National Essay Competition requires students to write an essay (up to 1,500 words) from the perspective of a medical professional.
There are essay titles available for Medicine, Dentistry , Veterinary Medicine and Allied Health .
For 2023, the essay questions are:
- Medicine – Should the patient be viewed as part of the multidisciplinary team?
- Dentistry – How can the holistic approach minimise periodontal disease in patients?
- Veterinary – What is the importance of a holistic approach when caring for livestock?
- Allied Heath – How can the multidisciplinary team optimise the care of the older person in hospital?
Deadline – midday on 1st May 2023.
Minds Underground Medicine Essay Competition
Minds Underground Medicine Essay Competition is aimed at students in Year 12, but younger students are also welcome to enter, and there are various essay title options to choose from. For the 2023 competition, one of the possible titles was ‘Should all healthcare be free? Discuss.’
The competition is designed to give students an opportunity to engage in research, hone their writing and argumentative skills, and prepare for university interviews. Minds Underground also runs essay competitions for other science subjects like Psychology and STEM.
The submission deadline is typically around March/April. Get more info here.
The Libra Essay Prize
The Libra Essay Prize is for students in Years 12 and 13 who are looking to prepare for university. Inspired by the admissions process at All Souls College, Oxford, entrants write an essay (1,500-2,000 words) responding to a single-word title.
For the 2023 prize, the single-word options were: Control, Collaboration, Exchange, Freedom, Claim.
Entrants are encouraged to use imagination in their essays to build interesting links between their chosen title and their school learning. There are prizes available of £50 for first place, £30 for second place and £20 for third place.
The deadline has varied from year-to-year: it was June in 2022 and April in 2023, so keep an eye on their website for more details.
Prep Packages
Get the best prep at the best price. Save up to 20% with prep packages for UCAT or Interview. Plus a new range of exciting medical experiences.
Trusted Courses
Our courses are trusted by world-leading schools and thousands of aspiring medics every year. 5*-rated UCAT and Interview Courses are available in person or live online.
1-2-1 Tutoring
Need one-to-one help? Our Doctor-trained Medicine tutors are ready to help you boost your UCAT score, nail your interviews and get you into Medical School.
Join Our Newsletter
Join our mailing list for weekly updates and tips on how to get into Medicine.
PREVIOUS ARTICLE
How To Set Up A Medical Society At School
NEXT ARTICLE
Medicine Entrance Exam Results: What's Next?
Loading More Content

medmentor ®
Year 12 essay competitions for medicine (updated 2022), essay competitions you can enter to make your medicine application stand out..

Why should I enter an essay competition?
One unique way of making your medicine application stand out is by entering various essay competitions that are relevant to science, technology, or healthcare.
By entering (and winning) essay competitions, medical schools will see that:
- You take an interest in medicine and education beyond what you're taught in your curriculum
- You're very proactive and dedicated
- You have excellent time management skills as you can juggle this with other academic commitments
- You're interested in writing and research
You don't need to win the competition for this to look good in your application. The fact that you've gone through all the effort to apply is fantastic in itself. It's something you can write about in your personal statement and bring up in your interviews. It's also something your teachers can write on your UCAS reference too.
It's important to remember there are many other important skills admission tutors andinterviewers will be looking for during your application. Our Medmentor SuperPack is tailor-made to help track your progress and plug any gaps to ensure you're the holistic candidate that every medical school is looking for.
Essay competitions for Year 12 students applying to medicine
Throughout the year, there are different essay competitions open for sixth form students to enter. We've compiled a list of some of the ones that are most relevant for your medicine application. Some of the deadlines are soon, whilst others are in a few months, so be selective and make sure you're giving yourself enough time to write something that is of a high standard.
American Society of Human Genetics Annual DNA Day Essay Contest
https://www.ashg.org/dna-day/ - Deadline: 2nd March 2022
You're expected to write a 750 word essay answering the following questions:
- How do Mendel’s discoveries help us understand Mendelian disorders?
- How does the study of Mendelian disorders help us understand complex diseases?
Please note essays must be submitted by a teacher or administrator however if you are home schooled parent submission is acceptable.
Newnham College University of Cambridge Medicine Essay Prize
https://newn.cam.ac.uk/wp-content/uploads/2022/01/Medicine-Information-and-Questions-2022.pdf - Deadline: 11th March 2022 at 12pm
Open to all female students currently in Year 12 at a UK state school. Choose 1 question from:
- How realistic is it to develop a small molecule therapy for Covid-19? Could such a therapy be rolled out in a timeframe that it could have an impact on the current pandemic?
- Sleep deprivation in clinical health settings. Does it matter?
- Looking to the future. Will stem cell therapies be outpaced by machine-brain interfaces for the treatment of retinal disease?
Minds Underground Medicine Competition
https://www.mindsunderground.com/medicine-competition - Deadline: 31st March 2022
Choose 1 question from:
- If you could invent a new drug, what would it be and why?
- What will the impact of an ageing population on the NHS look like?
- "This idea must die: We can't find new antimicrobials fast enough to make a difference." Do you agree?
Minds Underground Psychology Competition
www.mindsunderground.com/psychology-competition - Deadline: 31st March 2022
- What is more important: nature or nurture?
- Psychology Challenge: Design a research study.
- Does consciousness exist – how can we test for it?
Minds Underground Advanced Sciences Competition
www.mindsunderground.com/sciences-competition - Deadline: 31st March 2022
The questions you can choose from cover the different STEM subjects. Therefore, it may be preferable to select the one most closely related to medicine:
- Some people have argued that the discovery of DNA was the greatest scientific discovery of the 20th Century. Do you agree?
The Libra Education Essay Prize
www.libraeducation.co.uk/essay-prize - Deadline: 24th June 2022
You have a lot more flexibility and autonomy with this competition, where you can choose any essay title inspired by the following words.

The words that are arguably most relevant to medicine, science, and healthcare include 'sustainability', 'memory', and 'senses', though you can definitely make links with any of the words. For example, somebody could choose to write an essay on "Why is sustainability important in healthcare?" or "How has the pandemic impacted healthcare sustainability ?"
For these types of essays, think boldly and be unique if you want your essay to stand out.
Essay competitions for Year 12 students that will open later this year
Various other competitions will open up throughout the year. Keep tabs on these two pages below as their competitions will open up later this year.
- Robinson College Essay Prize (University of Cambridge) - https://www.robinson.cam.ac.uk/prospective-students/essay-prize
- University of Oxford Sixth-form prize in Medicine - https://www.ndorms.ox.ac.uk/research/research-groups/oxford-trauma/university-of-oxford-sixth-form-prize-in-medicine
How important are essay competitions for Year 12 students?
We would only recommend going out of your way to participate in essay competitions if you're quite comfortable with your academic grade. To write a good essay, you'll need to do extra reading and research in the topic area, and this can take some time. If your time is better spent revising and securing your grades, then please prioritise this. Essays are just another way to help your medicine application stand out, but they are by no means essential.
If you do have the time for it and you're not compromising your other commitments, then you've got nothing to lose - so give it your best shot!
Can I do anything else besides enter an essay competition?
If you're not quite ready to commit to an essay, you can also consider submitting articles to particular magazines or news outlets. If you're lucky, your piece may feature on their magazine, and this is sure to impress medical schools.
These don't have to be related to medicine or healthcare, it can be about your life as a student and any challenges you're experiencing. Even though it may not directly relate to medicine, it's certainly impressive and will show the medical school that you're not afraid to put yourself out there. For example:
- Consider writing for the Llama magazine: https://www.llamamag.com/about-the-llama/
- Push yourself further and submit a pitch to the Guardian https://www.theguardian.com/education/mortarboard/2013/sep/23/blogging-students-how-to-pitch-and-blog
Discover more golden opportunities to help you stand out in our Blog , Navigator and/or Superhub.
Recommended articles: How to Become a COVID-19 Vaccinator , Summer School Opportunities for Medical School Applications & Online Courses for Medical School Applicants
Prepare for Success with the SuperPack
Woolf Essay Prize 2024
The Woolf Essay Prize 2024 has now closed. Check back here in January 2025 for the 2025 competition!

In 1928, Virginia Woolf addressed the Newnham Arts Society on the Subject of ‘Women and Fiction’, and from this talk emerged her seminal text, A Room of One’s Own . Newnham is very proud of its place in the history of women’s education, and we are delighted in the continuation of the Woolf Essay Prize. A Room of One’s Own raises a number of questions surrounding the place of women in society, culture, and education, and the competition allows students to contemplate these themes and ideas while developing the independent research and writing skills essential to university-level study.
This year, the Woolf Essay Prize is open to all Women in Year 12 (or equivalent), regardless of school or country. For more information, including the question list, word limit, and submission details, please consult the Information and Questions document. The deadline for submission is 09:00am BST on Monday 8th July 2024. For any queries not answered here, please contact [email protected].
The Woolf Essay Prize will run separately to our Essay Writing Masterclass Programme , which encompasses a variety of subject interests.
This prize may be of particular interest to those studying English Literature, History, Politics, Philosophy or Sociology, but we absolutely welcome entries from interested students studying any combination of subjects.
Find out more about studying English Literature at University of Cambridge
Meet Shakespeare scholar Dr Bonnie Lander-Johnson
Please note information submitted as part of this competition will be used by the College for the purposes of assessing your essay as part of the Woolf Essay Prize. Certain personal details (questions 24-33 in the submission form) may also inform research which includes evaluation of the effectiveness of this programme for different types of participant, and equalities monitoring. All essay submissions are retained by the College permanently in the College Archive, with winning entries additionally published on the Newnham College website. If you have any questions about the use of your data, please contact [email protected] in the first instance.
Information about how your personal information will be used by us in connection with the administration of this event/activity, and for related purposes, is available here .
Fall 2024 Admissions is officially OPEN. Sign up for the next live information session here .
Discourse, debate, and analysis
Cambridge re:think essay competition 2024.
This year, CCIR saw over 4,200 submissions from more than 50 countries. Of these 4,200 essays, our jury panel, consists of scholars across the Atlantic, selected approximately 350 Honourable Mention students, and 33 award winners.
The mission of the Re:think essay competition has always been to encourage critical thinking and exploration of a wide range of thought-provoking and often controversial topics. The hope is to create a discourse capable of broadening our collective understanding and generating innovative solutions to contemporary challenges. This year’s submissions more than exceeded our expectations in terms of their depth and their critical engagement with the proposed topics. The decision process was, accordingly, difficult. After four rigorous rounds of blind review by scholars from Cambridge, Oxford, Stanford, MIT and several Ivy League universities, we have arrived at the following list of award recipients:
Competition Opens: 15th January, 2024
Essay Submission Deadline: 10th May, 2024 Result Announcement: 20th June, 2024 Award Ceremony and Dinner at the University of Cambridge: 30th July, 2024
We welcome talented high school students from diverse educational settings worldwide to contribute their unique perspectives to the competition.
Entry to the competition is free.
About the Competition
The spirit of the Re:think essay competition is to encourage critical thinking and exploration of a wide range of thought-provoking and often controversial topics. The competition covers a diverse array of subjects, from historical and present issues to speculative future scenarios. Participants are invited to engage deeply with these topics, critically analysing their various facets and implications. It promotes intellectual exploration and encourages participants to challenge established norms and beliefs, presenting opportunities to envision alternative futures, consider the consequences of new technologies, and reevaluate longstanding traditions.
Ultimately, our aim is to create a platform for students and scholars to share their perspectives on pressing issues of the past and future, with the hope of broadening our collective understanding and generating innovative solutions to contemporary challenges. This year’s competition aims to underscore the importance of discourse, debate, and critical analysis in addressing complex societal issues in nine areas, including:
Religion and Politics
Political science and law, linguistics, environment, sociology and philosophy, business and investment, public health and sustainability, biotechonology.
Artificial Intelligence
Neuroengineering
2024 essay prompts.
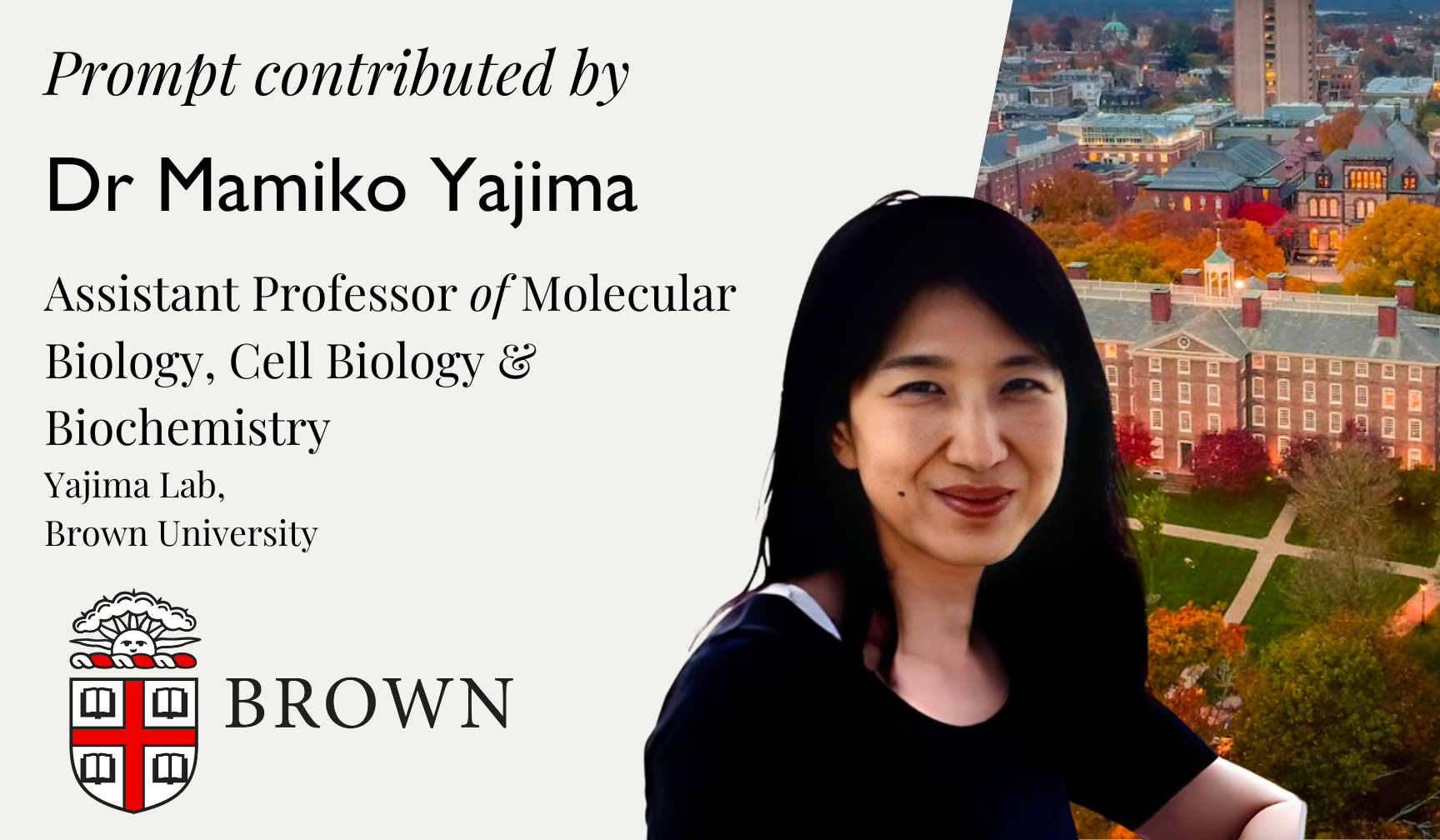
This year, the essay prompts are contributed by distinguished professors from Harvard, Brown, UC Berkeley, Cambridge, Oxford, and MIT.
Essay Guidelines and Judging Criteria
Review general guidelines, format guidelines, eligibility, judging criteria.
Awards and Award Ceremony
Award winners will be invited to attend the Award Ceremony and Dinner hosted at the King’s College, University of Cambridge. The Dinner is free of charge for select award recipients.
Registration and Submission
Register a participant account today and submit your essay before the deadline.
Advisory Committee and Judging Panel
The Cambridge Re:think Essay Competition is guided by an esteemed Advisory Committee comprising distinguished academics and experts from elite universities worldwide. These committee members, drawn from prestigious institutions, such as Harvard, Cambridge, Oxford, and MIT, bring diverse expertise in various disciplines.
They play a pivotal role in shaping the competition, contributing their insights to curate the themes and framework. Their collective knowledge and scholarly guidance ensure the competition’s relevance, academic rigour, and intellectual depth, setting the stage for aspiring minds to engage with thought-provoking topics and ideas.
We are honoured to invite the following distinguished professors to contribute to this year’s competition.
The judging panel of the competition comprises leading researchers and professors from Harvard, MIT, Stanford, Cambridge, and Oxford, engaging in a strictly double blind review process.

Keynote Speeches by 10 Nobel Laureates
We are beyond excited to announce that multiple Nobel laureates have confirmed to attend and speak at this year’s ceremony on 30th July, 2024 .
They will each be delivering a keynote speech to the attendees. Some of them distinguished speakers will speak virtually, while others will attend and present in person and attend the Reception at Cambridge.

The Official List of Re:Think 2024 Winners
Gold Recipients
- Ishan Amirthalingam, Anglo Chinese School (Independent), Singapore, Singapore
- Arnav Aphale, King Edward VI Camp Hill School for Boys Birmingham, West Midlands, United Kingdom
- Anchen Che, Shanghai Pinghe School, Shanghai China
- Chloe Huang, Westminster School, London, United Kingdom
- Rose Kim, MPW Cambridge, Cambridge, United Kingdom
- Jingyuan Li, St. Mark’s School, Southborough, United States
- Michael Noh, Korea International School, Pangyo Campus, Seoul, Korea
- Aarav Rastogi, Oberoi International School JVLR Campus, Mumbai, India
- Yuseon Song, Hickory Christian Academy, Hickory, United States
- Aiqi Yan, Basis International School Guangzhou, Guangzhou, China
Silver Recipients
- John Liu, Deerfield Academy, Deerfield, United States
- Sophie Reason, The Cheltenham Ladies College, Birmingham, United Kingdom
- Peida Han, Nanjing Foreign Language School, Nanjing, China
- Thura Linn Htet, Kolej Tuanku Ja’afar School, Negeri Sembilan, Malaysia
- Steven Wang, Radley College, Headington, United Kingdom
- Rainier Liu, Knox Grammar School, Sydney, Australia
- Anupriya Nayak, Amity International School, Saket, New Delhi, India
- Ming Min Yang, The Beacon School, New York City, United States
- Anna Zhou, Shanghai YK Pao School, Shanghai, China
- Yuyang Cui, The Williston Northampton School, Easthampton,United States
Bronze Recipients
- Giulia Marinari, Churchdown School Academy, Gloucestershire, United Kingdom
- Christina Wang, International School of Beijing, Beijing, China
- Chuhao Guo, Shenzhen Middle School, Shenzhen, China
- Isla Clayton, King’s College School Wimbledon, London, United Kingdom
- Hanqiao Li, The Experimental High School Attached to Beijing Normal University, Beijing, China
- He Hua Yip, Raffles Institution, Singapore, Singapore
- Wang Chon Chan, Macau Puiching Middle School, Macau, Macau, China
- Evan Hou, Rancho Cucamonga High School, Rancho Cucamonga, United States
- Carson Park, Seoul International School, Seongnam-si, Korea
- Sophie Eastham, King George V Sixth Form College, Liverpool, United Kingdom
The Logos Prize for Best Argument
- Ellisha Yao, German Swiss International School Hong Kong, Mong Kok, Hong Kong, China
The Pathos Prize for Best Writing
Isabelle Cox-Garleanu, Mary Institute and Saint Louis Country Day School, Frontenac, United States
The Ethos Prize for Best Research
Garrick Tan, Harrow School, Harrow on the Hill, United Kingdom

Gene therapy is a medical approach that treats or prevents disease by correcting the underlying genetic problem. Is gene therapy better than traditional medicines? What are the pros and cons of using gene therapy as a medicine? Is gene therapy justifiable?
Especially after Covid-19 mRNA vaccines, gene therapy is getting more and more interesting approach to cure. That’s why that could be interesting to think about. I believe that students will enjoy and learn a lot while they are investigating this topic.
The Hall at King’s College, Cambridge
The Hall was designed by William Wilkins in the 1820s and is considered one of the most magnificent halls of its era. The first High Table dinner in the Hall was held in February 1828, and ever since then, the splendid Hall has been where members of the college eat and where formal dinners have been held for centuries.
The Award Ceremony and Dinner will be held in the Hall in the evening of 30th July, 2024.

Stretching out down to the River Cam, the Back Lawn has one of the most iconic backdrop of King’s College Chapel.
The early evening reception will be hosted on the Back Lawn with the iconic Chapel in the background (weather permitting).

King’s College Chapel
With construction started in 1446 by Henry VI and took over a century to build, King’s College Chapel is one of the most iconic buildings in the world, and is a splendid example of late Gothic architecture.
Attendees are also granted complimentary access to the King’s College Chapel before and during the event.
Confirmed Nobel Laureates

Dr Thomas R. Cech
The nobel prize in chemistry 1989 , for the discovery of catalytic properties of rna.
Thomas Robert Cech is an American chemist who shared the 1989 Nobel Prize in Chemistry with Sidney Altman, for their discovery of the catalytic properties of RNA. Cech discovered that RNA could itself cut strands of RNA, suggesting that life might have started as RNA. He found that RNA can not only transmit instructions, but also that it can speed up the necessary reactions.
He also studied telomeres, and his lab discovered an enzyme, TERT (telomerase reverse transcriptase), which is part of the process of restoring telomeres after they are shortened during cell division.
As president of Howard Hughes Medical Institute, he promoted science education, and he teaches an undergraduate chemistry course at the University of Colorado

Sir Richard J. Roberts
The nobel prize in medicine 1993 .
F or the discovery of split genes
During 1969–1972, Sir Richard J. Roberts did postdoctoral research at Harvard University before moving to Cold Spring Harbor Laboratory, where he was hired by James Dewey Watson, a co-discoverer of the structure of DNA and a fellow Nobel laureate. In this period he also visited the MRC Laboratory of Molecular Biology for the first time, working alongside Fred Sanger. In 1977, he published his discovery of RNA splicing. In 1992, he moved to New England Biolabs. The following year, he shared a Nobel Prize with his former colleague at Cold Spring Harbor Phillip Allen Sharp.
His discovery of the alternative splicing of genes, in particular, has had a profound impact on the study and applications of molecular biology. The realisation that individual genes could exist as separate, disconnected segments within longer strands of DNA first arose in his 1977 study of adenovirus, one of the viruses responsible for causing the common cold. Robert’s research in this field resulted in a fundamental shift in our understanding of genetics, and has led to the discovery of split genes in higher organisms, including human beings.

Dr Aaron Ciechanover
The nobel prize in chemistry 2004 .
F or the discovery of ubiquitin-mediated protein degradation
Aaron Ciechanover is one of Israel’s first Nobel Laureates in science, earning his Nobel Prize in 2004 for his work in ubiquitination. He is honored for playing a central role in the history of Israel and in the history of the Technion – Israel Institute of Technology.
Dr Ciechanover is currently a Technion Distinguished Research Professor in the Ruth and Bruce Rappaport Faculty of Medicine and Research Institute at the Technion. He is a member of the Israel Academy of Sciences and Humanities, the Pontifical Academy of Sciences, the National Academy of Sciences of Ukraine, the Russian Academy of Sciences and is a foreign associate of the United States National Academy of Sciences. In 2008, he was a visiting Distinguished Chair Professor at NCKU, Taiwan. As part of Shenzhen’s 13th Five-Year Plan funding research in emerging technologies and opening “Nobel laureate research labs”, in 2018 he opened the Ciechanover Institute of Precision and Regenerative Medicine at the Chinese University of Hong Kong, Shenzhen campus.

Dr Robert Lefkowitz
The nobel prize in chemistry 2012 .
F or the discovery of G protein-coupled receptors
Robert Joseph Lefkowitz is an American physician (internist and cardiologist) and biochemist. He is best known for his discoveries that reveal the inner workings of an important family G protein-coupled receptors, for which he was awarded the 2012 Nobel Prize for Chemistry with Brian Kobilka. He is currently an Investigator with the Howard Hughes Medical Institute as well as a James B. Duke Professor of Medicine and Professor of Biochemistry and Chemistry at Duke University.
Dr Lefkowitz made a remarkable contribution in the mid-1980s when he and his colleagues cloned the gene first for the β-adrenergic receptor, and then rapidly thereafter, for a total of 8 adrenergic receptors (receptors for adrenaline and noradrenaline). This led to the seminal discovery that all GPCRs (which include the β-adrenergic receptor) have a very similar molecular structure. The structure is defined by an amino acid sequence which weaves its way back and forth across the plasma membrane seven times. Today we know that about 1,000 receptors in the human body belong to this same family. The importance of this is that all of these receptors use the same basic mechanisms so that pharmaceutical researchers now understand how to effectively target the largest receptor family in the human body. Today, as many as 30 to 50 percent of all prescription drugs are designed to “fit” like keys into the similarly structured locks of Dr Lefkowitz’ receptors—everything from anti-histamines to ulcer drugs to beta blockers that help relieve hypertension, angina and coronary disease.
Dr Lefkowitz is among the most highly cited researchers in the fields of biology, biochemistry, pharmacology, toxicology, and clinical medicine according to Thomson-ISI.
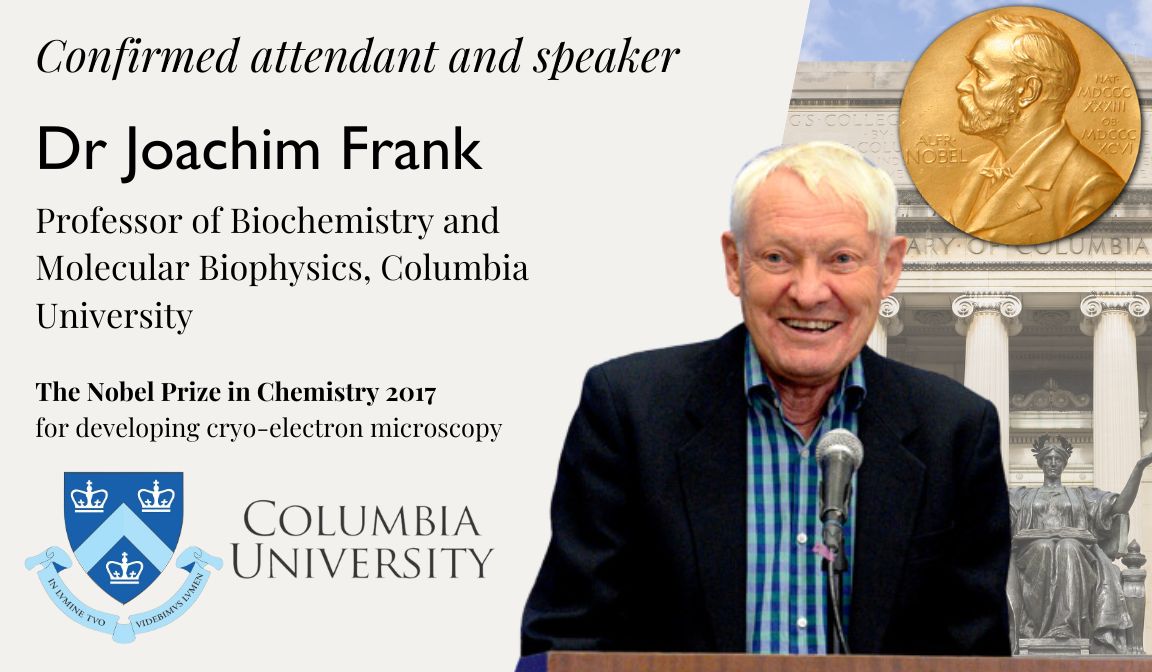
Dr Joachim Frank
The nobel prize in chemistry 2017 .
F or developing cryo-electron microscopy
Joachim Frank is a German-American biophysicist at Columbia University and a Nobel laureate. He is regarded as the founder of single-particle cryo-electron microscopy (cryo-EM), for which he shared the Nobel Prize in Chemistry in 2017 with Jacques Dubochet and Richard Henderson. He also made significant contributions to structure and function of the ribosome from bacteria and eukaryotes.
In 1975, Dr Frank was offered a position of senior research scientist in the Division of Laboratories and Research (now Wadsworth Center), New York State Department of Health,where he started working on single-particle approaches in electron microscopy. In 1985 he was appointed associate and then (1986) full professor at the newly formed Department of Biomedical Sciences of the University at Albany, State University of New York. In 1987 and 1994, he went on sabbaticals in Europe, one to work with Richard Henderson, Laboratory of Molecular Biology Medical Research Council in Cambridge and the other as a Humboldt Research Award winner with Kenneth C. Holmes, Max Planck Institute for Medical Research in Heidelberg. In 1998, Dr Frank was appointed investigator of the Howard Hughes Medical Institute (HHMI). Since 2003 he was also lecturer at Columbia University, and he joined Columbia University in 2008 as professor of Biochemistry and Molecular Biophysics and of biological sciences.

Dr Barry C. Barish
The nobel prize in physics 2017 .
For the decisive contributions to the detection of gravitational waves
Dr Barry Clark Barish is an American experimental physicist and Nobel Laureate. He is a Linde Professor of Physics, emeritus at California Institute of Technology and a leading expert on gravitational waves.
In 2017, Barish was awarded the Nobel Prize in Physics along with Rainer Weiss and Kip Thorne “for decisive contributions to the LIGO detector and the observation of gravitational waves”. He said, “I didn’t know if I would succeed. I was afraid I would fail, but because I tried, I had a breakthrough.”
In 2018, he joined the faculty at University of California, Riverside, becoming the university’s second Nobel Prize winner on the faculty.
In the fall of 2023, he joined Stony Brook University as the inaugural President’s Distinguished Endowed Chair in Physics.
In 2023, Dr Barish was awarded the National Medal of Science by President Biden in a White House ceremony.

Dr Harvey J. Alter
The nobel prize in medicine 2020 .
For the discovery of Hepatitis C virus
Dr Harvey J. Alter is an American medical researcher, virologist, physician and Nobel Prize laureate, who is best known for his work that led to the discovery of the hepatitis C virus. Alter is the former chief of the infectious disease section and the associate director for research of the Department of Transfusion Medicine at the Warren Grant Magnuson Clinical Center in the National Institutes of Health (NIH) in Bethesda, Maryland. In the mid-1970s, Alter and his research team demonstrated that most post-transfusion hepatitis cases were not due to hepatitis A or hepatitis B viruses. Working independently, Alter and Edward Tabor, a scientist at the U.S. Food and Drug Administration, proved through transmission studies in chimpanzees that a new form of hepatitis, initially called “non-A, non-B hepatitis” caused the infections, and that the causative agent was probably a virus. This work eventually led to the discovery of the hepatitis C virus in 1988, for which he shared the Nobel Prize in Physiology or Medicine in 2020 along with Michael Houghton and Charles M. Rice.
Dr Alter has received recognition for the research leading to the discovery of the virus that causes hepatitis C. He was awarded the Distinguished Service Medal, the highest award conferred to civilians in United States government public health service, and the 2000 Albert Lasker Award for Clinical Medical Research.

Dr Ardem Patapoutian
The nobel prize in medicine 2021 .
For discovering how pressure is translated into nerve impulses
Dr Ardem Patapoutian is an Lebanese-American molecular biologist, neuroscientist, and Nobel Prize laureate of Armenian descent. He is known for his work in characterising the PIEZO1, PIEZO2, and TRPM8 receptors that detect pressure, menthol, and temperature. Dr Patapoutian is a neuroscience professor and Howard Hughes Medical Institute investigator at Scripps Research in La Jolla, California. In 2021, he won the Nobel Prize in Physiology or Medicine jointly with David Julius.
Frequently Asked Questions
Why should I participate in the Re:think essay competition?
The Re:think Essay competition is meant to serve as fertile ground for honing writing skills, fostering critical thinking, and refining communication abilities. Winning or participating in reputable contests can lead to recognition, awards, scholarships, or even publication opportunities, elevating your academic profile for college applications and future endeavours. Moreover, these competitions facilitate intellectual growth by encouraging exploration of diverse topics, while also providing networking opportunities and exposure to peers, educators, and professionals. Beyond accolades, they instil confidence, prepare for higher education demands, and often allow you to contribute meaningfully to societal conversations or causes, making an impact with your ideas.
Who is eligible to enter the Re:think essay competition?
As long as you’re currently attending high school, regardless of your location or background, you’re eligible to participate. We welcome students from diverse educational settings worldwide to contribute their unique perspectives to the competition.
Is there any entry fee for the competition?
There is no entry fee for the competition. Waiving the entry fee for our essay competition demonstrates CCIR’s dedication to equity. CCIR believes everyone should have an equal chance to participate and showcase their talents, regardless of financial circumstances. Removing this barrier ensures a diverse pool of participants and emphasises merit and creativity over economic capacity, fostering a fair and inclusive environment for all contributors.
Subscribe for Competition Updates
If you are interested to receive latest information and updates of this year’s competition, please sign up here.
Carissa Lee
2nd Year Medical Student, Norwich Medical School
I am a second year medical student at Norwich Medical School with an interest in cardiology, preventative medicine and public health. I am really interested in using big data to optimise efficiency within the healthcare system and to provide more equitable access for all. When I’m not studying, I go for long runs, travel and have a good laugh with friends. I hope to work in a big city one day!
Farah Saleh Hijazi
Final Year Medical Student, Qatar University
I am a final year medical student at Qatar University interested in pursuing a career in Anesthesiology. I am passionate about merging my interests in medicine and graphic designing. Apart from medicine, I enjoy reading and crochet.

Robyn Wilcha
Fifth Year Medical Student, The University of Manchester
I am a fifth-year medical student studying at The University of Manchester with strong interests in psychiatry, medical education and research. Through the COVID-19 pandemic, I have worked as a data intern with Public Health England (PHE); this role has allowed me to appreciate the extent of the pandemic and has driven me to further understand the long-term implications from a medical and economic perspective.
Highly Commended
- Rasha Rashid
- Ishveer Sanghera
Secondary School Pupils

Ruairí Moore
A-Level Student, St. Michael's College, Enniskillen
I am currently studying Biology, Chemistry, Maths and Physics at A-level aspiring to study Medicine in the future. I am fascinated by the complexity of human anatomy and welcome the challenges that a career in Medicine would bring. I enjoy a range of sports including Gaelic Football, Hurling, and cross country, and I also play the drums in my free time.

Sasha Hammond
6th form student
I am looking forward to applying to medical school and currently plan on specializing in psychiatry, as this is a field with a large scope for research and the development of new treatments. Outside of my studies, I enjoy painting from nature and rowing.

Destiny-Alliah Colombo
Year 11 Student, Whitley Academy
I am a Year 11 student who is interested in studying Maths, Chemistry and Biology at a higher level. I have been inspired to enter this essay competition due to my interest in reading medicine at University, and I have enjoyed developing my writing and researching skills through the process of formulating this essay.
- Preesha Jain
- Seoyoon Kim

Apimaan Yogeswaran
Intercalation (5th Year), Norwich Medical School
Cardiology is set in my heart as the career I want to pursue. With the perfect balance between medicine, surgery, and patient contact, I feel it leaves many of the right doors open for me. Always fascinated with the human body, my intrigue further developed upon reading ‘Homo Deus’ which discussed the potential evolution of humanity through technology, in particular healthcare AI. Aside from medicine, I enjoy variety in my life – from dancing salsa, to cooking Asian cuisine, to running – I strongly believe that we shouldn’t let life opportunities pass us no matter how career-focused we are.

Ferenc Kohsuke Gutai
4th Year Medical Student, University of Dundee
I am a fourth-year medical student in the University of Dundee with strong interests in the neurosciences as well as the use of technology in medicine (including AI). I thoroughly enjoyed my BMedSci in Anatomy in Dundee and look forwardto exploring the field of surgery in the future. In my spare time I enjoy playing the violin and dabbling in philosophy, aviation and programming.

Aneirin Potter
King's College, London
I studied Neuroscience at King’s College Cambridge before working in the John Radcliffe Neurology ward and moving to graduate-entry medicine at King’s College, London. I am interested in neuroscience’s role in medicine and when/how science and academia inform people’s lives.
- Lucy Lazzereschi
- Marcus Powis
- Patrick Chan
- Rameez Naqvi
- Soham Bandyopadhyay

Sarah Mackel
Secondary School Pupil, International School of Luxembourg
I have a lifelong passion for learning, and recently completed a student internship at the Centre Hospitalier de Luxembourg as well as a course on neurodegenerative diseases at the Barcelona Center for Genomic Regulation. Myschool has the IB program, where I study Chemistry, Biology, and English Literature at HL, and look forward to applying to medical school. Aside from academics, I enjoy reading, playing field hockey, and composing various genres of music.

Aasiyah Faizal
Year 12, London Academy Of Excellence, Tottenham
I am a year 12 student who is interested in studying medicine as I have a genuine fascination in the core understanding of the human body and how it works. I also love the ever-advancing and evolving nature of medicine which makes it areally stimulating and engaging course as you are constantly learning. In my free time, I enjoy painting watercolour floral anatomy as well as reading and staying up to date about different topics in human geography.

Mehzabeen Hakim
Year 12, Chelmsford County High School for Girls
I am currently studying Biology, Chemistry, Maths and Psychology at A-Level, with hopes of reading Medicine at university in the future. In addition to this, I volunteer at a primary school and a children’s hospital, as well as partaking an active role in charity work within my school. Having recently been appointed as BioChemSoc president and alongside doing an EPQ project, it has inspired me to explore the world of academic research and enter myself into essay competitions.
- Angelique Chan
- Daneel Louw
- Estelle Elise Rollinger
- Manaal Menahood
- Yasmeen Rahali

Claire Read
Second year medical student at Nottingham University
I’m a keen runner and compete as part of the University distance running team but aside from my sport, I enjoy keeping myself busy; for example taking part in this essay competition. I also volunteer with primary school children, teaching them what to expect in a hospital which is really fun as I’m potentially interested in paediatrics or General practice as a career. Shadowing GP’s has been my favourite part of the course so far; I love chatting to patients which is a good change up from lectures!

Tobi Rotimi
Final year medical student at University College London
I am passionate about medicine because it brings together so many areas that I enjoy including research, patient centred care, life-long learning and teaching. I am heavily interested in Neurology, Paediatrics and Ear, Nose and Throat surgery. Hopefully I will have a career that combines medical education, research and clinical practice in one of these specialties in the future.

Ciara Doyle
Third year medical student at Warwick University
My previous degree was in Biomedical Science which encouraged me to build my portfolio by working in an NHS based lab for a year before applying to medicine. Recent experience in research projects has seen me undertake several studies both independently and as a team. This is something I look forward to building on in the future, specifically within women’s health, which is an area of medicine I am particularly passionate about.
- Ellie Phelps
- Chon Meng Lam
- Harriet Conley

Keunyoung Bae
Year 12 at Cheongshim International Academy, South Korea
I am interested in studying liberal arts and sciences together. I hope to study abroad in the United States or United Kingdom for university. My school follows the AP curriculum, and I am currently taking AP Biology, Chemistry, Literature, Calculus BC, Computer Science, and Comparative Politics. I wish to become a pathologist one day and research cancer with people around the world. I love to play the flute and listen to music in my free time.

Keza Nganga
Year 12 at Rugby School, UK
I have always been passionate about the sciences and am looking forward to applying to medical school in a couple of years. I really enjoy reading anything from novels to the national geographic and I’m always ready to read beyond the textbook. Away from the classroom I enjoy working in the design blocks designing something from scratch as well taking part in photography.

Kutloogh Qureshi
Year 12 at The Tiffin Girls’ School, UK
I am currently studying Biology, Chemistry, Maths and English Literature and hope to study medicine at university. What draws me to medicine is the ability to help those in need whilst contributing to a wider scientific community. I currently volunteer in an elderly care ward at my local hospital and in my spare time I like to do rowing, write poetry and read as much as I can.

Year 12 at North London Collegiate School, UK
I have a passion for Science, and am currently working through the Chemistry, Biology, Maths and Physics A Level syllabus’. I particularly enjoy doing practicals in school. I am committed to a wide range of extra-curricular activities including lacrosse, swimming and playing the violin and piano. Being a member of the school science blog has encouraged me to research into science outside of my examination specifications. I read for pleasure and to follow advances in Medicine. I find listening to music and spending time with my friends and pet rabbits relaxing.

Year 13 at Caterham School, UK
I am thoroughly interested in medicine due to the depth and complexity the career provides. The choices available in this career are near endless yet medical ethics will have a role in any speciality. It is essential we give some attention to the morals and principles of medicine which is why I chose this essay. In my free time, I enjoy playing badminton, watching movies and charity.
- Anna Armstrong
- Radek Antczak
- Sasha Westcott
- Sebastian Rensburg
- Misaki Okuyama
- Matilda Rose
- Marie Barberon
- Jessica Seddon
- Daniel Collier
- Millie Thomas
- Lisa George
- Katy Salvesen
- Meghana Vipin
- Nazifa Syed
- Deniz Tanritanir
- Rhea Burman
- Katherine King
- Nancy Braithwaite
- Ashitha Abdul
- Afzaa Altaf
- Jodie Ferris
- Bavisan Jeyakanthan
- Mathew John

Loukas Kouzaris
Third year graduate entry medical student at Swansea University
I am passionate about the sciences, in particular medicine, and enjoy the challenges of working within the modern-day NHS. My time spent studying medicine has fuelled my enthusiasm for teaching and has shown me the benefits of teamwork in a pressured system. I consider continued learning one of the factors that make medicine so rewarding. This ongoing development only encourages opportunities to teach your successors whilst providing a mutually beneficial facet to develop communication & interpersonal skills that can be taken into future practice.

Abhiyan Bhandari
Second year medical student at University College London
During my first year I have been involved in the role of an Advanced Student Academic Representative, liaising closely with the Medical School department heads to improve the delivery, structure and content of the first year modules. Also, as President of UCL Nepalese Society, I am able to demonstrate my leadership and management skills and work with committee members to organise and manage events and the society. As a member of the BMA, I am keen on updating my knowledge on the current affairs of the medical sector. In my spare time, I enjoy playing badminton and cooking as a way of keeping active and de-stressing. Having contributed as a student reviewer in the upcoming Orthopaedic X-Ray book, I am keen to contribute my skills in medical education. I am also currently actively involved in a research project involving T1 mapping of inflammation in cardiac MRI.

Omair Sadiq
Before starting medical school, I always had an interest in research and it was something I wanted to carry on doing whilst studying medicine. Due to time and workload pressures at medical school, carrying out lab research wasn’t very feasible so I decided to carry out research on paper rather than in labs. This is where I came across essay competitions and it is something I find very stimulating, challenging and very manageable. It provides me with an opportunity to express myself and carry out research which may be beneficial to someone somewhere.

11th grade student at L’Ecole Chempaka, Trivandrum, India
I’m fascinated by how the human body works and I plan to pursue a career in biomedical research in the future. Microbiology, Infectious Disease and Epidemiology are a few areas that have recently piqued my interest, which is why I decided to write my essay on antibiotic resistance. Apart from my academic interests, I love playing the flute and I’m also an avid debater and quizzer.

Kelly Ka Lee Li
Year 11 student at Wycombe Abbey School, UK
As an aspiring medical doctor, I strive towards opportunities in order to supplement my knowledge beyond the standard syllabuses. I also enjoy researching a variety of fields within medicine and the sciences in general, as well as playing the piano and the violin in my spare time.

Harry Geddes
Year 12 student at Kings College Taunton, UK
I am currently studying Biology, Chemistry and Maths at A-level. I am currently volunteering in a care home and this has helped me to understand some of the reoccurring issues in modern medicine that I have talked about in my essay. In my spare time I like to play football, hockey and cricket as well as playing the piano and the clarinet.

Juliet Makkison
Year 12 student at Brookfield Community School, UK
I’m studying Biology, Chemistry, Maths and Further Maths currently and I would like to study medicine at University. In my free time I’m a young leader at a Brownie Group and help out at local care home.
- Simar Bajaj
- Eden Simkins
- Zain Girach
- Erica Ogbomo
- Evie Singer
- Shivani Raja
- Vindhya Maripuri
- Rahul Panniker
- Shivani Pedda Venkatagari
- Kirthika Janaarthanan
- Lucy Norman
- Lauren Davies
- Aydah Sarah Ashraf
- Neha Telikapalli
- Anthony Kemp
- Amy Turnbull
- Reika Yamamoto

Richard Odle
5th year MB/PhD student at the University of Cambridge
I am in my research phase at the Babraham Institute whilst continuing medical education. My current ambition is to become a clinician-scientist, taking the best of both worlds! I have a particular interest in the development of novel cancer therapeutics.


Yusuf Karmali
Intercalating in Experimental Pathology at Barts and The London
Having completed three years of medicine. I have interests in both medical education and health policy, hence why this essay was something that intrigued me!

Michael Smith
First year medical student at the University of Warwick
In July 2016 I graduated with a B.Sc in Mathematics from Southampton, and I enjoyed education so much, that within 2 months I was back in full time lectures. Although there is quite a notable change from Mathematics to Medicine, and I can’t wait to see what the next 4 years have in store! (Although I feel that hoping there is some time off in there too, might be setting my hopes too high)

Corey Magee
Fourth year medical student at Queen’s University, Belfast
I am currently taking time away from medicine to complete an intercalated masters program. While this is certainly proving to be a challenging endeavour, I am enjoying the research exposure that is on offer. In terms of my future career, I have yet to decide on one particular speciality. In fact, I tend to enjoy all parts of the medical course…well, nearly all of them! Alongside medicine, my other thriving passions include music and drama!

Lucy Faulkner
Final year medical student at the University of Sheffield
I have completed a Bmedsci in the department of Infection and Immunity and in the future I hope to complete paediatrics training and then specialise in clinical genetics, after really enjoying my elective in the clinical genetics department of Sheffield Children’s’ Hospital as well as paediatrics on the Isle of Man. I’m looking forward to starting the Academic Foundation Programme in Yorkshire in August this year. In my spare time I am a member of the University of Sheffield Triathalon club.

Chai Chung Sien
Second year medical student in the University of Dundee
I enjoy participating in activities both inside and outside the medical school and have helped out in a few events organised by the Dundee Malaysian Society (DUMAS). I have a special interest in Human Factors and Surgery and look forward to doing a project integrating the two. Apart from academics, I especially enjoy listening to the oldies, reading fantasy novels and playing sports.

Ashvin Kuri
Studying Biology, Chemistry and Mathematics at A-Level
I hope to study Medicine at University. I particularly enjoy Human Biology and learning about diseases and their cures. Outside of lessons, I enjoy competing in Athletics, playing the violin, piano and singing, and volunteering in my local area at a Care Home and with disabled children.

Nabihah Rahman
Studying Biology, Chemistry, Maths and Latin at A-level
I intend to pursue a career in medicine in the future and specialise in Paediatrics. I am employed as a pharmacy sales assistant and I also enjoy volunteering at my local hospital and care home.
Aleksandra Skroban
Well done to Aleksandra on being a runner-up in the secondary school category of our ‘Unofficial Guide to Medicine Essay Competition’ for her essay on “Doctor accountability and ‘no-blame’ culture as instruments to reduce ‘never events’ “

Poppy Pierce
Studying Chemistry, Biology, Physics and Psychology at Haggerston Sixth form
I want to study medicine so that I can care for those who are at their most vulnerable. I love the satisfaction of helping people and I know medical school will give me the skills to do so. I believe there is so much more research still to be done. In medicine you see the rewards of your labour unfold before your eyes and the field of expertise continually expands.

Molly Teather
Studying Biology, Chemistry, Maths and French A levels
I am hoping to study Medicine at university and become a doctor. I chose this title after having shadowed an anaesthetist which has given me a particular interest in anaesthetics and surgery.

Studying Chemistry, Biology, Physics and Mathematics at AS Level
I would like to study Medicine at university and specialise in Infectious Diseases. Outside of school I enjoy playing the piano and volunteering at my local hospital and church.

Andrea McCarthy
University College Dublin
My name is Andrea Mc Carthy and I am a final year student in University College Dublin. I enjoy working with my hands so after first training as a physiotherapist, I returned to school to study medicine and am hoping to specialise in surgery. Which kind? Well, that’s a work in progress but I do like general surgery and orthopaedics best. When I am not freaking out about becoming an intern or trying to get into yet another surgery, I enjoy shooting and catching up with friends. Thanks for reading and enjoy!

Eloise Shaw
Third year medical student, University of Nottingham
Having just completed the BMedSci part of her degree Ellie is looking forward to beginning the clinical phase, starting working on the wards and hopefully finding the field of medicine she wants to specialise in.
Michaela Gaston
Reigate Grammar School

Akua Crankson
James Allen’s Girls’ School
My name is Akua Crankson and I am 17 years old. I live in London and am currently studying Biology, Maths, Chemistry and Latin at A2 level. I hope to study medicine at university and specialise in Geriatrics. In my spare time I enjoy playing the clarinet and volunteering at my local hospital.

Mehnaz Khan
Ibstock Place School
My name is Mehnaz Khan. I am 17 years old and am currently studying mathematics, chemistry and biology at a level. I absolutely love biology, learning about the human body and about all the complex yet fascinating wonders of medical science. One day, I hope to become a doctor and work to make people’s lives better.

Vinay Mandagere
Stockport Grammar School
Hello, I’m Vinay. I’m a Gap Year student who is entering the University of Bristol this year to study medicine. I enjoy reading, playing the violin and trying different types of tea (especially Earl Grey). I’m also very interested in writing.
Are you a medical trainee, a student, a doctor or medical professional?
We’d love to hear from you for feedback or if you would like to contribute towards the Unoffical Guide to Medicine series.
- Zeshan Qureshi Award for Outstanding Achievement in Medical Education
- Getting Involved
Pin It on Pinterest
By continuing to use the site, you agree to the use of cookies. more information Accept
The cookie settings on this website are set to "allow cookies" to give you the best browsing experience possible. If you continue to use this website without changing your cookie settings or you click "Accept" below then you are consenting to this.

Prizes for students
The RSM has numerous prizes and awards aimed at students. By submitting an application, you will have the chance to present at a meeting, add to your CV and open up more career opportunities.
Please note this page is updated regularly.
All submissions must be sent in by 11.59pm on the stated deadline date.
Generally prizes, awards or travel grants must be claimed before the end of the academic year in which they are gained (30 September).
For more information or help please contact [email protected]
General Practice with Primary Healthcare
John Fry prize
Deadline: Thursday 1 August 2024
Open to: Medical, nursing and allied healthcare students with an interest in general practice and primary care
CAIPE John Horder Team Award and John Horder Student Award
Deadline: Wednesday 31 July 2024
Open to: Individuals or teams working within the community who can demonstrate outstanding principles of collaborative working and the Student annual essay award offered to a student who has been involved with interprofessional learning or working, within the community.
Team Award prize: Certificate of achievement and £600, Student Award prize: Certificate of achievement and £150
History of Medicine Society
History of Medicine Society: Sarah Hughes Trust Prize in conjunction with the Medical Journalist Association
Deadline: Sunday 1 September 2024
Open to: Any emerging or established journalist and/ or healthcare practitioner who has written a piece or made a broadcast that debunks fake news in the field of medical journalism over the last 12 months.
Prize: £1000
Obstetrics & Gynaecology
Dame Josephine Barnes Award
Deadline: Wednesday 21 August 2024 at 11:59pm
Open to: All medical undergraduate students
Prize: £100
Patient Safety
Patient Safety Section: Oral presentation
Deadline: Thursday 5 September 2024 at 23:59
Open to: All students
Prize: A year's free RSM membership Apply here
Patient Safety Section: Poster presentation
Respiratory Medicine
Student Award
Submission deadline: Monday 3 February 2025
Open to: All current UK medical students and FY1 doctors who completed the research whilst at medical school.
Prize: £100 and free attendance to the RSM Respiratory Careers Day on Monday 12 May 2025
Surgery section: MIA Prize
Deadline : Sunday 22 September 2024
Open to : Surgical trainees, pre-doctoral surgical researchers and students.
Surgery Section: Adrian Tanner Prize
Deadline : Sunday 22 September 2024
Open to : All surgical trainees and foundation doctors (Medical students should apply for the John Dawson prize)
Surgery Section: John Dawson Medical Student Prize
Deadline : Sunday 22 September 2024
Open to : Medical students (Qualified doctors should apply for the Adrian Tanner Prize)
Vascular, Lipid & Metabolic Medicine
The Harvey prize
Submission deadline: Thursday 15 August 2024
Open to: Trainees and students
Oral presentation prize: £150 plus one year's RSM membership
Poster presentation prize: £100
"It’s a great privilege for our team to have been recognised by this prestigious award from the RSM Surgery Section and reflects the dedicated work behind this exciting innovation."
See more from prizes and awards or hear from previous prize winners.
- About the CMJ
- Editorial Board
- Submission Guidelines
- Submit Your Article
- Manage Submission

A Decade of Progress in Global Neurosurgery
- Perspectives
- Read more about A Decade of Progress in Global Neurosurgery
- Log in to post comments
COVID-19, Care and Chaos: A Medical Student’s Experience of the Intensive Care Unit
Janvi Karia shares her reflections of working in ICU as a 5th year medical student during the COVID-19 pandemic with the CMJ.
- Read more about COVID-19, Care and Chaos: A Medical Student’s Experience of the Intensive Care Unit
What impact can simulations have in advancing medicine?
- Read more about What impact can simulations have in advancing medicine?
What advancements in clinical neurosciences need to occur in the next 10 years?

- Read more about What advancements in clinical neurosciences need to occur in the next 10 years?
The Ethics of Curtailing Paediatric Services in the Face of the COVID-19 Pandemic
- Read more about The Ethics of Curtailing Paediatric Services in the Face of the COVID-19 Pandemic
Group consultations - the future of General Practice?

- Read more about Group consultations - the future of General Practice?
Human papillomavirus: the case for a gender-neutral vaccination programme
Human papillomaviruses (HPV) are a large family of double-stranded DNA viruses that infect skin and mucosal cells [1] . Of the more than 100 recognised genotypes, at least 13 are considered to be oncogenic (or ‘high-risk’). The two most common of these, i.e. type 16 and 18, are known to cause approximately 70 per cent of all cervical cancers. Oncogenic HPVs have also been implicated as the aetiological agents in squamous cell carcinoma of the anus, genitals and the head and neck.
- Read more about Human papillomavirus: the case for a gender-neutral vaccination programme
The Dawn of Precision Medicine in Stroke
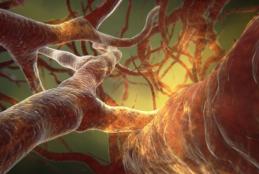
- Read more about The Dawn of Precision Medicine in Stroke
Management of severe shock in a young adult following lower GI bleeding within a rural context
- Read more about Management of severe shock in a young adult following lower GI bleeding within a rural context
Biodiversity and the importance of environmental literacy in medical education
Doctors have an important role in the protection and promotion of human health, from individual to global scales. As our climate changes and ecosystems are degraded, the protection of nature is becoming increasingly important for public and global health efforts. There is an increasingly large body of evidence highlighting the environment and human health as critically connected – the variety of ecosystems on Earth provide human populations with many necessities for health and wellbeing.
- Read more about Biodiversity and the importance of environmental literacy in medical education
- Editorial Team

Copyright © Cambridge Medical Journal | ISSN 2046-1798
Cambridge Essay Competitions
Essay competitions are brilliant for a number of reasons!
You can use them to:
The essay competitions usually become open for submissions after the winter holidays. Be sure to check any eligibility criteria, requirements and deadlines. This page will be updated when new competitions are announced, and when deadlines are passed, so check back regularly! All essay competitions and events at Cambridge (both online and in-person) can be found here 🔗 🌟.
Magdalene College Arts and Humanities Essay Competition 2024 🔗 🌟 Any student in their penultimate year at a state school can enter this competition, which will open in early 2024. Last year, there were 12 questions covering a variety of topics within the arms and humanities - you can read the winning entries here 🔗. To register your interest in this competition for 2024, fill in this form 🔗.
Fitzwilliam College Essay Competitions: various subjects 🔗 🌟 Fitzwilliam College runs a variety of competitions in Ancient World and Classics, Archaeology, History, Land Economy, Medieval World, Architecture, and Economics (this last one is for state-school UK students only). All competitions are open to Year 12 students and are designed to encourage students to pursue their interests in subjects they might not be able to study in depth at school. Last year, the deadline for all competition entries was the 1st of March, so make sure to check back in early 2024 for updates.
Newnham College Woolf Essay Prize 🔗 🌟 The Woolf Essay Competition is focussed on women in literature, history, society and culture. There are also competitions for other subjects - more information these will be coming soon. Webinars to help support your entry can be found here 🔗 . The deadline for the Woolf Prize last year was the 14th of July.
Girton College Humanities Writing Competition 🔗 An opportunity for students interested in pursuing any humanities subject to write creatively! Year 12 students may enter with an essay or piece of creative writing using an object from Girton College’s small antiquities museum, the Lawrence Room, as their prompt.
Robinson College Essay Prize: various subjects 🔗 Year 12 students may submit an essay of up to 2,000 words in response to one of the set questions, which cover a wide variety of academic subjects. The prize did not run in 2023, but may in 2024.
Trinity College Essay Prizes 🔗 These competitions give entrants the opportunity to write an essay of up to 2,000 words in response to the set question/(s). Last year there were competitions for English Literature, Launguages, Linguistics, Philosophy, Politics, Law, and History.
Did you spot a typo or formatting issue? Let us know by emailing us at [email protected] .
The following pages contain information about our Essay Prizes run for Lower and Upper 6th Students internationally, including how to apply.
The Robson History Prize will not run in 2024 but we are expecting to run it again in 2025.
Gould Prize for Essays in English Literature
Languages and Cultures Essay Prize
Linguistics Essay Prize
Philosophy Essay Prize
R.A. Butler Politics Prize
Robert Walker Prize for Essays in Law
- Share on Twitter
- Share on Facebook
- Share on LinkedIn
- Share via email
- previous post: Information for Offer Holders
- next post: Gould prize for essays in English Literature
Access and Outreach Hub
Privacy overview.
| Cookie | Duration | Description |
|---|---|---|
| __cf_bm | 30 minutes | This cookie, set by Cloudflare, is used to support Cloudflare Bot Management. |
| S | 1 hour | Used by Yahoo to provide ads, content or analytics. |
| Cookie | Duration | Description |
|---|---|---|
| AWSALB | 7 days | AWSALB is an application load balancer cookie set by Amazon Web Services to map the session to the target. |
| AWSELBCORS | 1 hour | This cookie is used by Elastic Load Balancing from Amazon Web Services to effectively balance load on the servers. |
| Cookie | Duration | Description |
|---|---|---|
| _cb | never | This cookie stores a visitor's unique identifier for Chartbeat tracking on the site. |
| _cb_svref | never | Chartbeat sets this cookie to store the original referrer for the site visitor. This cookie expires after 30 minutes but the timer is reset if the visitor visits a new page on the site before the cookie expires. |
| _chartbeat2 | never | Chartbeat sets this cookie to store information about when a visitor has visited the site before. This helps to distinguish between new, returning, and loyal visitors. |
| CONSENT | 2 years | YouTube sets this cookie via embedded youtube-videos and registers anonymous statistical data. |
| iutk | 5 months 27 days | This cookie is used by Issuu analytic system to gather information regarding visitor activity on Issuu products. |
| Cookie | Duration | Description |
|---|---|---|
| mc | 1 year 1 month | Quantserve sets the mc cookie to anonymously track user behaviour on the website. |
| NID | 6 months | NID cookie, set by Google, is used for advertising purposes; to limit the number of times the user sees an ad, to mute unwanted ads, and to measure the effectiveness of ads. |
| VISITOR_INFO1_LIVE | 5 months 27 days | A cookie set by YouTube to measure bandwidth that determines whether the user gets the new or old player interface. |
| YSC | session | YSC cookie is set by Youtube and is used to track the views of embedded videos on Youtube pages. |
| yt-remote-connected-devices | never | YouTube sets this cookie to store the video preferences of the user using embedded YouTube video. |
| yt-remote-device-id | never | YouTube sets this cookie to store the video preferences of the user using embedded YouTube video. |
| yt.innertube::nextId | never | This cookie, set by YouTube, registers a unique ID to store data on what videos from YouTube the user has seen. |
| yt.innertube::requests | never | This cookie, set by YouTube, registers a unique ID to store data on what videos from YouTube the user has seen. |
| Cookie | Duration | Description |
|---|---|---|
| __Host-JSESSIONID | session | No description |
| __wpdm_client | session | No description |
| COMPASS | 1 hour | No description |
| DotMetrics.DeviceKey | 1 year | This cookie is set by the provider Dotmetrics.net. This cookie is used for implementation of the technical part of the MOSS measurement. |
| DotMetrics.UniqueUserIdentityCookie | 1 year | This cookie is set by the provider Dotmetrics.net. This cookie is used for implementation of the technical part of the MOSS measurement. |
| MONITOR_WEB_ID | 3 months | No description available. |
| NSC_QH-PMY | session | No description available. |
| ttwid | 1 year | No description available. |
| X-Mapping-ckfcdfng | session | No description |
Medicine related essay competitions
Quick Reply
Related discussions.
- What a levels should I choose for law?
- LSE: advice for EXTRAcurriculars and Personal statement
- International Relations/Politics/History Extracurriculars
- Applying to PPE at Oxford Tips?
- Medicine or law
- medicine essay competitions
- what does cambridge actually want from a medical applicant?
- stem extra curriculars/work experience (year 10)
- 2023 Immerse education essay competition
- Will maths help with Cambridge Medicine
- Great, I want to get into medicine. Now what?
- Immerse Education essay competition 2022
- DPsych Counselling psychology GCU - 2024
- Advice on applying to medicine
- How many essay competitions/supercurriculars can I join? Any recs?
- 4 A Levels and EPQ - should I drop the EPQ
- volunteering for politics and international relations
- 4 A levels and WBQ
- Wider Research & Activities for Medicine Application
- Which universities would be realistic?
Last reply 26 minutes ago
Last reply 50 minutes ago
Last reply 10 hours ago
Last reply 1 day ago
Last reply 3 days ago
Last reply 4 days ago
Last reply 1 week ago
Last reply 2 weeks ago
Last reply 3 weeks ago
Articles for you

A guide to the Oxbridge application process

Bmat: when it is, what it costs and how to prepare

Personal statement FAQs

The basics of applying to university: glossary of terms
Omsk State Medical University Russia 2024-25: Fees, Ranking, Admission, Courses, Eligibility etc.

Omsk State Medical University, located in Omsk, Siberia, is a top choice for studying MBBS in Russia. Founded in 1920 as the Medical Faculty of the Siberian Institute of Veterinary Medicine and Zoology, it offers a variety of undergraduate and postgraduate programs in different medical fields, including General Medicine, Pediatrics, Dentistry, Pharmacy, and Preventive Medicine.
| Abroad University MBBS Application Form 2024 | |
|---|---|
| Manipal Pokhara College of Medical Science, Pokhara, Nepal | |
| Kursk State Medical University, Russia | |
| Grigol Robakidze University, Georgia | |
This article provides information about Omsk State Medical University, mainly for Indian students, such as Faculties, courses offered, admission process, eligibility criteria, fee structure, Ranking in Russia etc.
[Page Index]
College summary.
Before we complete the college summary, let us look at the major details of Omsk State Medical University Russia .
Wants to Study MBBS Abroad from a top Country with low tution Fees? Subscribe Now!
| Omsk State Medical University Russia | |
| OSMU Russia | |
| Omsk Oblast, Russia | |
| 1920 | |
| Public | |
| Maria A. Livzan | |
| English & Russian | |
| Not Required | |
| , | |
| Ministry of Health of the Russian Federation | |
| MBBS | |
| USD: $ 3,700/- (Annually) INR: Rs. 3,07,100/- (Annually) | |
| Country- 89 World- 4042 ( : Edurank) | |
| September Intake | |
| Yes (Male & Female) | |
| Omsk Tsentralny Airport | |
| http://omsk-osma.ru/ | |
Affiliation and Recognition
The Omsk State Medical University is one of the biggest medical universities in Russia, and it is affiliated and recognized by various Medical Councils such as:
- National Medical Commission of India (NMC).
- Ministry of Science and Higher Education of the Russian Federation.
- World Health Organization (WHO).
Omsk State Medical University Consists of five faculties. Here we have listed below:
- Faculty of General Medicine
- Faculty of Pediatrics
- Faculty of Dentistry
- Faculty of Preventive Medicine
- Faculty of Pharmacy
Courses Offered
Omsk State Medical University Russia Courses offer quality medical programs under highly qualified faculty and state-of-the-art infrastructure. It is famous for its undergraduate medical programs.

| MBBS | 6 Years (English Medium) 7 Years (Russian Medium) |
Why study at the Omsk State Medical University Russia?
- The medium of instruction at the University is English.
- There are good dormitory facilities for local as well as international students.
- Omsk has creative groups for regional, national and international competitions and thus is a world of opportunities and activities for students.
- Omsk State University has 22 specialized sports clubs, including five fitness centres, two large gyms and an Olympic sports complex with a pool.
Admission Procedure
If you want to take Omsk State Medical University admission to Russia in 2024-25, you must qualify for the National Eligibility Entrance Exam (NEET) for Indian students.
Eligibility Criteria
In this section, all the students check the eligibility criteria of Omsk State Medical University Russia.
| Your age should be at least 17 years old on or before 31st December of the admission year. *No Upper Age Limit. | |
| Class 12th in Science, with PCB and English subjects from a board recognized by the authorities in India. | |
| 60% in 10+2 (UR) 50% (SC/OBC/ST) | |
| (For Indian Students) |
Graphical Representation of Eligibility Criteria
Documents Required
Before admission to Omsk State Medical University Russia, please do not forget to carry all these related documents.
- Passport (Minimum 18 months validity).
- 10th Certificate & Mark sheet.
- 12th Certificate & Mark sheet.
- Birth Certificate.
- 10 passport-size Photographs.
- Official Invitation letter from the Medical University of Russia.
- Authorization of all documents from the Ministry of External Affairs, New Delhi.
- Legalization of all documents from the Russian Embassy.
- Bank receipt of 1st year of Omsk State Medical University MBBS fees (required for some universities).
- HIV test documents.
Fee Structure 2024-25
In this section, all the MBBS students get information about the Omsk State Medical University MBBS fees in 2024. Check all the relevant queries regarding fees following this page: Low fees for MBBS Colleges in Russia .
| Tuition Fee | $ 3,700/- | Rs. 3,07,100/- |
| Hostel Fee | $ 700/- | Rs. 58,100/- |
| Total Fee | $ 4,400/- | Rs. 3,65,200/- |
Ranking 2024-25
According to Edurank, the Omsk State Medical University ranking in Russia and all over the world ranking:
| Country Ranking | 89 |
| World Ranking | 4042 |
Advantages of MBBS in Russia
About Omsk City
- Omsk City is located on the Irtysh River in the southwestern Siberia region. It is Russia’s seventh-largest city.
- In 1782, Omsk was granted town status.
- The main and important economy of the city involves agriculture and the retail sectors.
- Omsk City has a humid continental climate based on the constantly changing weather swing. Omsk experiences more than 300 sunny days in a year.
- Major road, rail, and air hub pathways connect Omsk. Metro, trolleybuses, trams, and shared taxis are also available in the city of Omsk.
- Some of the city’s main attractions are the Omsk Drama Theatre, Green Island, Central Vrubel Museum of Fine Arts, etc.
- The city’s famous foods are Okroshka, Shanxi, Pelmeni, Kurnik, Mors, Bird Cherry cake, etc.
Temperature
Contact Details
Omsk State Medical University Russia (OSMU Russia) Address: Ulitsa Lenina, 12, Omsk, Omsk Oblast, Russia, 644099 Contact Number : +91 7827-804-741
Omsk State Medical University Photos

Frequently Asked Questions (FAQs)
When is summer vacation starting at omsk state medical university russia.
The summer vacation starts from 1st July to 31st August at the Omsk State Medical University.
What type of food is available at the hostel?
Indian food is available at the hostel. All types of vegetarian and non-vegetarian food are available at the hostel, and in Russia, there is a wide variety of milk products and fruits.
How is the classroom environment at Omsk State Medical University Russia?
At Omsk State University, the teacher-student ratio of 1:10 gives students a more attentive and personal learning environment.
Which facilities does the university provide for student’s physical fitness?
The university provides well-equipped gyms, sports and recreation centres, and physical fitness ski centres.
- Admission Updates,
- Top Collages
You may like to read
- LN Medical College Kyrgyzstan 2024-25: Admission, Courses, Fees, Ranking etc.
- International Higher School of Medicine Kyrgyzstan 2024-25: Fees, Ranking, Course, Admission etc.
- Kyrgyz State Medical Academy Kyrgyzstan 2024-25: Admission, Courses, Fees, Ranking, Eligibility etc.
- Devdaha Medical College and Research Institute 2024-25: Admission, Courses, Fees, Ranking etc.
About Harsh
Hello, I'm Harsh Begwani, with a year of expertise in MBBS and Ayush courses. I have detailed knowledge of various colleges' fee structures, cutoffs, and intake procedures. If you're looking for insights or assistance in pursuing MBBS or BAMS courses, feel free to comment below—I'm here to help!
Leave a Comment Cancel reply
Notify me via e-mail if anyone answers my comment.
Abroad MBBS Update 2024 : Admission Dates, Top College, Fees, Location, Scholarship etc.
Get admission to Top Overseas Universities with Affordable Fees.
Breadcrumbs Section. Click here to navigate to respective pages.

Omsk Oblast
DOI link for Omsk Oblast
Click here to navigate to parent product.
This chapter presents history, economic statistics, and federal government directories of Omsk Oblast. Omsk Oblast is situated in the south of the Western Siberian Plain on the middle reaches of the Irtysh river. Kazakhstan lies to the south. Tyumen Oblast lies to the north-west, and Tomsk Oblast and Novosibirsk Oblast lie to the east. The city of Omsk was founded as a fortress in 1716. In 1918 it became the seat of Adm. Aleksandr Kolchak's 'white' 'All-Russian Government'. Omsk fell to the Bolsheviks in 1919, and Kolchak 'abdicated' in January 1920. In 2015 Omsk Oblast's gross regional product (GRP) amounted to 617,184m. roubles, equivalent to 311,973 roubles per head. The Oblast's soil is the fertile black earth characteristic of the region. Its agriculture consists mainly of animal husbandry, hunting and the production of grain. The sector employed 14.6% of the workforce and contributed 9.6% of GRP in 2015.
- Privacy Policy
- Terms & Conditions
- Cookie Policy
- Taylor & Francis Online
- Taylor & Francis Group
- Students/Researchers
- Librarians/Institutions
Connect with us
Registered in England & Wales No. 3099067 5 Howick Place | London | SW1P 1WG © 2023 Informa UK Limited
- Open access
- Published: 25 July 2019
Deciphering drug resistance in Mycobacterium tuberculosis using whole-genome sequencing: progress, promise, and challenges
- Keira A. Cohen 1 ,
- Abigail L. Manson 2 ,
- Christopher A. Desjardins 2 ,
- Thomas Abeel 2 , 3 &
- Ashlee M. Earl ORCID: orcid.org/0000-0001-7857-9145 2
Genome Medicine volume 11 , Article number: 45 ( 2019 ) Cite this article
17k Accesses
87 Citations
10 Altmetric
Metrics details
Tuberculosis (TB) is a global infectious threat that is intensified by an increasing incidence of highly drug-resistant disease. Whole-genome sequencing (WGS) studies of Mycobacterium tuberculosis , the causative agent of TB, have greatly increased our understanding of this pathogen. Since the first M. tuberculosis genome was published in 1998, WGS has provided a more complete account of the genomic features that cause resistance in populations of M. tuberculosis , has helped to fill gaps in our knowledge of how both classical and new antitubercular drugs work, and has identified specific mutations that allow M. tuberculosis to escape the effects of these drugs. WGS studies have also revealed how resistance evolves both within an individual patient and within patient populations, including the important roles of de novo acquisition of resistance and clonal spread. These findings have informed decisions about which drug-resistance mutations should be included on extended diagnostic panels. From its origins as a basic science technique, WGS of M. tuberculosis is becoming part of the modern clinical microbiology laboratory, promising rapid and improved detection of drug resistance, and detailed and real-time epidemiology of TB outbreaks. We review the successes and highlight the challenges that remain in applying WGS to improve the control of drug-resistant TB through monitoring its evolution and spread, and to inform more rapid and effective diagnostic and therapeutic strategies.
Mycobacterium tuberculosis is the causative agent of tuberculosis (TB), which is most often spread person-to-person via cough aerosols. Although many individuals who are exposed to M. tuberculosis never develop active disease, the World Health Organization (WHO) estimated 10 million new cases of active TB and 1.3 million deaths in 2017 alone [ 1 ].
Since its initial documentation in the 1940s [ 2 ], drug-resistant TB has threatened public health control efforts. In 2016, there were an estimated 490,000 new cases of multidrug-resistant (MDR) TB, which is defined by phenotypic resistance to both isoniazid and rifampicin [ 3 ]. Approximately 10% of MDR-TB cases globally can be classified as extensively drug-resistant (XDR), indicating that there is concomitant resistance to quinolones (such as the fluoroquinolones, levofloxacin, and moxifloxacin) and to a second-line injectable agent (amikacin, kanamycin, or capreomycin) [ 3 ]. As expected, drug-resistance patterns predict treatment outcome; in 2015, TB treatment success overall was 83%, whereas the success rate was 54% for MDR-TB or rifampicin-resistant-TB (RR-TB) and only 30% for XDR-TB [ 4 ].
Culture-based techniques remain the current reference standard for both diagnosis and drug-susceptibility testing of TB, but these processes are time-consuming and require specialized laboratory capacity. More recently, the use of rapid molecular tests for the diagnosis of TB has increased globally, particularly the use of Xpert MTB/RIF (Cepheid, Sunnyvale, CA), a PCR-based assay that simultaneously detects the presence of M. tuberculosis and resistance to rifampicin.
Current recommendations for the treatment of drug-susceptible TB include a 6-month course of a multi-drug regimen of rifampicin, isoniazid, pyrazinamide, and ethambutol. Historically, treatment of MDR- or XDR-TB involved the long-term use of second-line drugs, including injectable agents [ 5 ]. More recently, the MDR-TB treatment landscape has changed with the introduction of multiple novel second-line drugs that can be administered orally (Table 1 ). In 2012, bedaquiline, a diarylquinolone, became the first TB drug from a novel drug class to receive US Food and Drug Administration (FDA) approval in over 40 years [ 48 , 49 ] (Table 1 ). Another oral agent, delamanid, a nitro-dihydro-imidazooxazole derivative, has also shown promise for TB treatment [ 50 , 51 ].
In 2018, the WHO published updated treatment guidelines for MDR/RR-TB [ 47 ], recommending fully oral MDR regimens for many patient groups. Recommended treatment strategies include both shorter, standardized MDR regimens (for 9 to 12 months) and longer, individualized treatment regimens (for 18 to 20 months). The updated guidelines group antitubercular drugs on the basis of how they should be combined to create individualized, longer MDR-TB regimens [ 47 ] (Table 1 ).
Despite advances in both diagnostics and therapeutics for TB, challenges remain. Obstacles for rapid M. tuberculosis diagnosis include: (i) the imperfect sensitivity of molecular tests for the detection of this pathogen, particularly in the case of paucibacillary TB (where there is a lower bacterial burden); (ii) lack of comprehensive molecular assays due to incomplete knowledge of all resistance mutations in TB; and (iii) technical limitations to the numbers of mutations that can be included on diagnostic molecular platforms. Furthermore, the rollout of rapid diagnostic platforms to low-resource settings has been a challenge. Remaining treatment challenges include: prolonged treatment courses, leading to greater drug exposure, toxicity, and non-compliance; unacceptable side-effect profiles; logistics of drug access; and re-infection [ 52 ].
The dawning of the new age of genome sequencing began to revolutionize our approach to human diseases, including TB. In 1998, Cole et al. [ 53 ] reported the complete genome sequence of the M. tuberculosis reference strain H37Rv, which was approximately 4.41 million base pairs in length and encoded approximately 4000 genes. The first sequencing of a clinical reference strain, CDC1551, quickly followed [ 54 ]. An accompanying editorial optimistically stated: “After several decades in the slow lane of classical microbiology, M. tuberculosis is once again at the cutting edge of science” [ 55 ]. However, even at the time of these breakthroughs, there was recognition that translating these genomic data into clinical benefit would prove challenging [ 55 ]. Despite these challenges, it is clear, more than 20 years later, that M. tuberculosis genomic data have been remarkably useful in improving our understanding of how drug-resistant TB evolves and spreads and in helping to inform diagnostics and therapies.
In this review, we discuss the molecular epidemiologic and diagnostic advances made by sequencing M. tuberculosis , with a focus on drug-resistant TB. We do not review the practice of whole-genome sequencing (WGS) of M. tuberculosis as this has been reviewed recently [ 56 ]. Key findings that are discussed include the use of WGS to identify drug-resistance determinants in M. tuberculosis and to elucidate the evolution and spread of drug-resistant organisms, and the clinical applications of this technology (Table 2 ).
Identifying M. tuberculosis drug-resistance determinants
Drug resistance in M. tuberculosis is the result of chromosomal mutations in existing genes that are passed along through vertical descent, that is, passed from mother to daughter cells. Unlike many other bacterial pathogens, M. tuberculosis rarely recombines via lateral exchange of DNA [ 83 ] and also lacks plasmids. Many of the resistance determinants were discovered before the sequencing of the M. tuberculosis genome was completed. By 1998, resistance mechanisms had already been discovered for classical first- and second-line TB drugs including isoniazid (alterations in genes katG and inhA ); rifampicin (in rpoB ); streptomycin (in rrs and rpsL ); pyrazinamide (in pncA ); ethambutol (in embB ); quinolones (in gyrA ); and kanamycin (in rrs ) (reviewed in Ramaswamy and Musser [ 84 ]) (Table 1 ). However, the targeted amplification and sequencing of known or suspected resistance genes revealed that these mechanisms were insufficient to explain all phenotypic resistance [ 85 , 86 ], and resistance mechanisms for several newer drugs—including pretomanid, bedaquiline, and delamanid—were discovered over the next eight years during a period when WGS was becoming routine. Together, in the past 20 years, WGS-based approaches, focused on both laboratory-derived and naturally circulating populations of drug-resistant M. tuberculosis , have provided a more complete account of the genomic features that cause treatment resistance, enabling the identification of novel resistance mechanisms for existing drugs, and the determination of the mechanisms of action of newly discovered drugs.
Identifying resistance determinants in laboratory-derived mutants
Drug-resistant mutants can be derived in vitro by growing drug-susceptible M. tuberculosis strains in drug-containing media, and selecting for mutants that are able to grow in the presence of the drug. Sequencing laboratory-derived resistant mutants has played a critical role in identifying both the mechanism of action of new TB drug classes, including diarylquinolines (for example, bedaquiline) [ 19 ] and nitroimidazopyrans (for example, PA-824) [ 19 , 29 ], and rare resistance mechanisms for established antitubercular drugs, including ethambutol [ 15 ], pyrazinamide [ 13 ], carbapenems [ 30 ], cycloserine [ 87 ], clofazimine, and bedaquiline [ 20 ]. For example, WGS of laboratory mutants identified drug efflux as a mechanism of resistance to clofazimine and bedaquiline [ 20 , 21 , 22 ], and this approach continues to be a mainstay for identifying the mechanism of action of compounds that are in development for TB [ 88 ].
Although laboratory-derived mutants are helpful in elucidating novel resistance mechanisms, mutations that have evolved in laboratory settings do not always match those in clinical isolates of drug-resistant M. tuberculosis [ 89 , 90 ], for reasons that are largely unknown. Studies by Ford et al. [ 91 , 92 ] suggested that these mismatches could not be explained by differences in the mutation rate in these settings, because the in vitro mutation rate of M. tuberculosis correlates well with the in vivo mutation rate in both humans and in a macaque model. Differences in the relative fitness of specific mutants grown in in vitro compared to in vivo conditions are probably responsible for these mismatches, but more work is needed. Regardless of the reason, if the goal is to identify a full complement of resistance mutations on which to base molecular diagnostics, isolates from clinical collections must be studied as these bacteria have evolved their resistance within the host.
Quantifying and identifying resistance determinants in clinical strains
Among the larger studies exploring resistance in natural populations, Walker et al. [ 58 ] analyzed the genomes of 3651 drug-resistant and -susceptible M. tuberculosis isolates for associations between phenotypic resistance to eight first- and second-line drugs, and then predicted genotypic resistance on the basis of a compiled catalog of 232 resistance mutations in 23 candidate resistance genes. Resistance to most drugs could be predicted accurately, with a mean sensitivity of 92% and specificity of 98%, suggesting that the majority of resistance—particularly for first-line drugs—is explained by known mechanisms and mutations (Table 1 ). Numerous other studies have found similar results using smaller datasets [ 7 , 25 , 57 , 69 , 93 , 94 ]. This result was echoed in a more recent study by the Comprehensive Resistance Prediction for Tuberculosis (CRYPTIC) Consortium and the 100,000 Genomes Project that focused solely on first-line drugs, which included analysis of 10,209 globally diverse M. tuberculosis isolate genomes against a database of mutations identified in a literature search [ 60 ]. Notably, predictions for mutations that are associated with resistance to pyrazinamide were greatly improved over earlier predictions; this study achieved 91.3% sensitivity in predicting resistance to this drug compared to 57% sensitivity in Walker et al. [ 58 ]. Although the news has been encouraging with respect to completing the catalog of mutations that cause resistance to first-line drugs, few studies have attempted to predict resistance to second-line drugs [ 95 ]. Some of these drugs, such as D-cycloserine, pyrazinamide, and para-aminosalicylic acid (PAS), are more difficult to assay because they have been reported to have variable drug phenotypes in clinical microbiology laboratories [ 96 ] (discussed later).
To fill gaps in the catalog of drug-resistance mechanisms, genome-wide association study (GWAS) approaches, originally designed for use on human genomic data, have been adapted for non-recombining microbes such as M. tuberculosis and used to predict novel resistance mechanisms [ 97 , 98 ] (Table 3 ). The majority of GWAS predictions remain experimentally unverified, but several new resistance-associated genotypes have been validated. Farhat et al. [ 7 ] sequenced 116 M. tuberculosis genomes and developed a phylogenetic convergence test, ‘PhyC’, to identify resistance associations. They identified a mutation in ponA1 (c.1095G>T) and showed that it conferred a minimum inhibitory concentration (MIC) to rifampicin that was twofold higher than that of wild-type bacteria. Zhang et al. [ 57 ] sequenced 161 genomes from China and searched for genes that appeared to be under positive selection and more frequently mutated in drug-resistant isolates. Resistance-associated polymorphisms in two intergenic regions upstream of the known resistance genes thyA -Rv2765 and thyX - hsdS .1 were shown to cause increased gene expression of a lacZ construct in Mycobacterium smegmatis , suggesting that these mutations may mediate PAS resistance through the overexpression of downstream genes.
Desjardins et al. [ 25 ] used a combination of the correlated evolution test [ 104 ] (to test for correlated evolution of genotype and phenotype) and a simple GWAS framework to search for novel drug-resistance mechanisms in 498 genomes from South Africa and China. Of note, they combined all variants within each gene that were predicted to inactivate gene function, and used these combinations as the input into the association test to increase statistical power in the detection of genomic features that are associated with resistance. They found that loss-of-function mutations in ald (Rv2780), which is predicted to encode an alanine dehydrogenase, correlated with unexplained resistance [ 25 ]. They also confirmed experimentally that these mutations conferred increased resistance of laboratory and clinical isolates to D-cycloserine [ 25 ], a key drug in MDR- and XDR-TB regimens that has severe psychiatric and central nervous system toxicities.
Hicks et al. [ 105 ] used the algorithm phyOverlap to perform a GWAS on 549 clinical isolates from China, in which they identified mutations that disproportionately occurred in isoniazid-resistant isolates. In addition to known resistance and compensatory mutations for first- and second-line drugs, they identified an association with prpR (Rv1129c). They then went on to characterize prpR as a transcriptional regulator of propionate metabolism which, instead of drug resistance, confers tolerance to multiple antibiotics in a macrophage model of infection.
In one of the largest GWAS published to date, Coll et al. [ 59 ] combined PhyC with a GWAS approach within a mixed-regression framework to detect determinants of resistance to 14 drugs in a large dataset of 6465 global M. tuberculosis clinical isolates. Although no functional experiments were performed to validate the predictions, new resistance-associated mutations were identified, including new codons in ethA (a gene known to activate ethionamide, which is a prodrug) that are associated with ethionamide resistance, and mutations in the thyX promoter associated with PAS resistance. Mutations in the promoter of thyX have been previously shown to upregulate thyX [ 43 , 57 , 106 ].
Predicting susceptibility and drug resistance in M. tuberculosis
As the list of suspected resistance determinants grows, there has been a need to establish well-curated databases of drug-resistance single nucleotide polymorphisms (SNPs) [ 107 ]. Initially, SNP databases, including TBDB [ 108 ] and PATRIC [ 109 ], were created to bring together genome annotation data and other functional data. Unfortunately, some of the pioneering databases of drug-resistance-associated mutations in M. tuberculosis , including TBDReamDB [ 110 ], have not been maintained to include emerging data.
Software and web-based tools have also been developed to enable the community to infer TB drug resistance from WGS data. These tools include CASTB [ 111 ], KVarQ [ 112 ], MyKrobe Predictor TB [ 113 ], PhyResSE [ 114 ], TBProfiler [ 115 ], and TGS-TB [ 116 ]. Studies have compared the sensitivity and specificity of these tools in predicting drug resistance [ 117 , 118 , 119 ], and have found that they tend to perform quite well for first-line drugs but less optimally for second-line drugs. In addition to tools, there have been improvements to databases, including the creation of the Relational Sequencing TB Database Platform (ReSeqTB) [ 120 , 121 ] and efforts from the CRyPTIC Consortium [ 122 ], which seeks to develop a curated database of clinically relevant drug-resistance mutations.
Continued refinement of these drug-resistance databases and prediction tools is necessary. Miotto et al. [ 123 ] performed a systematic review in which they assigned a confidence level to associations of individual and groups of mutations with phenotypic drug resistance. Importantly, they identified that certain mutations that are included in current commercial diagnostic tests, including eis c-2a, do not have a convincing association with drug resistance. Input from ongoing large sequencing projects will be needed to optimize the inference of resistance phenotypes from sequence data, especially for mutations that are present at low frequency in natural populations.
Challenges in uncovering the remaining resistance elements
Although WGS approaches have been successful in identifying resistance mechanisms, there are computational and experimental challenges that hamper efforts to complete the catalog of TB drug resistance. For example, for non-recombining organisms such as M. tuberculosis , interpretation of GWAS output can be complicated because non-causal variation can be tightly linked to causal variation [ 124 ]. Furthermore, as a result of frequent multidrug resistance, resistance mutations for one drug can appear to be highly associated with phenotypic resistance to multiple drugs [ 25 ], and confirmatory wet lab studies, which are non-trivial in M. tuberculosis , are often necessary to identify causal resistance mutations correctly. In addition, genotype–phenotype associations are largely dependent on accurate phylogenies, and phylogenetic reconstruction can be challenging in M. tuberculosis because of its slow rate of evolution [ 92 , 125 , 126 , 127 , 128 ], which gives rise to relatively few SNPs in clinical isolates.
When defining phenotypic resistance, different studies often use different drug concentration cutoffs and test in different media, complicating the meta-analysis of multiple datasets. In addition, phenotypic resistance testing of some antitubercular drugs, including pyrazinamide and D-cycloserine, is notoriously challenging and unreliable [ 129 ], introducing phenotypic inaccuracies that can confound analyses. Furthermore, the dichotomous classification of phenotypic resistance as ‘resistant’ or ‘susceptible’ will fail to identify drug-resistance mutations that result only in minimal increases in MIC, and there is emerging evidence that such mutations may be clinically relevant. TB relapse following treatment has been found to occur more commonly in individuals who harbored M. tuberculosis isolates that were susceptible to, yet had minimally increased MIC values for, either isoniazid or rifampicin [ 130 ]. Future study designs that address phenotypic resistance as a spectrum, rather than a binary value, will be needed to identify such mutations.
Heteroresistance, defined as the coexistence of pathogen populations that have differing nucleotides at a specific drug-resistance locus [ 131 ], can also confound genotype–phenotype comparisons [ 132 , 133 , 134 ]. A bacterial culture in which only a small fraction of the population is resistant can appear to be resistant when tested on media containing a drug, yet when grown on drug-free media for genome sequencing, the sensitive fraction can dominate, resulting in a genotypic prediction of sensitivity [ 132 ]. The problem of heteroresistance seems to be particularly common with fluoroquinolone resistance [ 135 ].
Last, innate characteristics of the M. tuberculosis genome—namely, highly repetitive DNA sequences and the high guanine-cytosine (GC) content of the genome (65.6%) [ 53 ]—present technical difficulties for both WGS and bioinformatic analyses. GC-rich regions can be troublesome for library PCR amplification and sequencing, and reads that represent highly repetitive regions of the genome can confound alignments by mapping to multiple regions of the genome and hampering accurate de novo assemblies. In addition, approximately 10% of the coding regions in M. tuberculosis are dedicated to two repetitive protein families that are unique to mycobacteria (the PE and PPE families), which have conserved Pro-Glu (PE) and Pro-Pro-Glu (PPE) motifs [ 53 ]. Even with WGS investigation [ 136 ], the function of the PE and PPE genes has remained elusive, although recent studies have suggested that they may play a role in virulence [ 137 ]. Their association with drug resistance remains largely unexplored because bioinformatic studies of M. tuberculosis often exclude these genes [ 138 , 139 ]. In the future, long-read sequencing technology may allow these regions to be sequenced successfully in order to assess if they have a role in drug resistance.
Understanding the evolution and spread of drug resistance in M. tuberculosis
Prior to WGS, the diversity and epidemiology of resistant M. tuberculosis were examined using DNA fingerprinting techniques, like IS6110 restriction fragment length polymorphism (RFLP) typing [ 140 ], spoligotyping (spacer oligonucleotide typing, a method of typing strains according to the distinct hybridization patterns of their spacer oligonucleotides) [ 141 ], and mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU-VNTR) typing [ 142 , 143 , 144 , 145 ]. These techniques enabled assessments of the diversity of resistant strains in specific geographic regions [ 146 , 147 , 148 , 149 ] and, when combined with the genetic profiling of resistance mutations, allowed strain-level monitoring of patients on TB therapy [ 150 ].
The dramatic increase in resolution afforded by WGS has extended the sensitivity and resolution with which the diversity and evolution of drug-resistant M. tuberculosis can be assessed. This has resulted in the more confident identification of cases of recent transmission [ 151 ] and re-infection [ 152 ], and has provided insights into the evolution of resistance within individual patients and across populations. WGS has also enabled more sensitive differentiation of de novo acquisition of resistance (where resistance mutations emerge within a host) from person-to-person transmission of resistance, a critical capability given that these two scenarios require different health-system responses in order to stem resistance.
Within patient evolution of drug resistance
Despite the slow evolutionary rate of M. tuberculosis , estimated at 0.3–0.6 SNPs/genome/year [ 69 , 92 , 125 , 126 , 127 , 128 ], experimental data suggest that drug resistance can evolve within an individual patient during TB treatment. Eldholm et al. [ 61 ] described the first documented case of XDR evolution of M. tuberculosis from a fully susceptible ancestor within a single patient, by sequencing nine serial isolates collected over a 42-month period. During this time, seven known resistance mutations emerged in a stepwise fashion after the clinical use of each corresponding drug, revealing how TB drug pressures can rapidly shape M. tuberculosis populations in vivo.
However, the evolution of drug resistance within a host is not always linear, and instead can involve a complex interplay of heterogeneous M. tuberculosis populations [ 153 , 154 ]. In particular, transient genetic diversity can exist before a dominant clone emerges. In addition, as the size of the transmission bottleneck (the number of bacteria transmitted during an infection event) in M. tuberculosis is not well understood [ 155 ], it is difficult to estimate the relative contribution of diversity that is transmitted to the patient versus diversity that evolves within the patient. Numerous WGS studies, performed either on isolates or directly on DNA extracted from serially collected sputum samples, have revealed substantial transient genetic diversity in pathogen populations within patients, particularly within resistance genes [ 61 , 62 , 106 , 156 , 157 , 158 , 159 ]. This diversity was observed to endure months before a single variant became fixed in the population (the situation when only a single variant remains). In the study by Eldholm et al. [ 61 ] mentioned above, the seven resistance-conferring mutations that eventually dominated were from amongst 35 mutations observed in total throughout the sampling period [ 61 , 160 ]. They joined eight other mutations that were not resistance-associated but that also became fixed in the population, probably as the result of a phenomenon called ‘hitchhiking’ in which non-adaptive mutations are selected because of their linkage and physical proximity to consequential mutations.
The relative fitness cost of drug-resistance mutations often determines which mutations become fixed within a host. While multiple mutations that confer resistance to a specific drug can evolve repeatedly, mutations conferring no or little fitness cost are typically selected, resulting in fixed dominant mutations [ 61 , 156 ]. Compensatory mutations (discussed in more detail later), which serve to counterbalance the deleterious effects of acquired resistance, have also been shown to emerge during treatment [ 156 ].
WGS has also revealed how combination chemotherapy effectively prevents the emergence of drug resistance during treatment for TB. In a study of very deep WGS of serial sputum specimens from patients receiving treatment for TB, Trauner et al. [ 62 ] demonstrated that the combined action of multiple active drugs prevented transient mutants from fixing within a population and becoming dominant. The fewer the drugs that were applied, the more likely it was that resistance would develop and become fixed.
Population views of drug-resistance evolution
A number of careful WGS studies have empirically established SNP-based criteria to discriminate cases of recent transmission from unrelated infections—usually using the criterion that recently transmitted strains differ by fewer than 6–12 total SNPs across the M. tuberculosis genome [ 63 , 125 , 126 , 161 ]. In a 2016 review, Nikolayevskyy and colleagues [ 63 ] systematically compared WGS to fingerprinting techniques for detecting transmission, including a meta-analysis of 12 studies published between 2005 and 2014. They concluded that results from WGS studies not only have higher discriminatory power, but they also enable more sensitive detection of transmission events that may have been missed by epidemiologic methods.
Although traditional spoligotyping analyses suggested that drug-resistant strains were diverse, WGS of clinical isolates began to reveal the full breadth of diversity in resistant M. tuberculosis . The TB epidemic in South Africa over the past two decades has been well-studied in this regard. In an early WGS investigation, Ioerger et al. [ 64 ] examined 14 phenotypically diverse strains within the Beijing lineage and showed that resistance mutations arose independently multiple times, and that XDR isolates may be less fit and less able to transmit. WGS studies across larger sets of strains from the same region in South Africa suggested that, although de novo resistance is indeed common, highly resistant strains (including MDR and XDR strains) have the ability to spread broadly by person-to-person transmission. This includes the ongoing transmission of a circulating XDR clone in South Africa that is linked to the infamous Tugela Ferry XDR outbreak [ 162 ] that brought XDR-TB to the world stage in 2005. A more recent large-scale study confirmed that XDR strains have been broadly transmitted person-to-person in KwaZulu-Natal [ 65 ].
The patterns observed in South Africa hold for many other parts of the world. Recent studies have shown that patterns of both de novo evolution and person-to-person spread of drug resistance in M. tuberculosis also occur in Belarus, Russia, England, and Malawi [ 73 , 139 , 159 , 163 , 164 ]. In a composite analysis of over 5000 M. tuberculosis isolates from patients from around the globe, Manson et al. [ 66 ] confirmed that both de novo evolution and person-to-person transmission are important factors for the rise and spread of drug-resistant TB worldwide. The emergence of MDR and XDR M. tuberculosis was found to be a frequent occurrence that is distributed fairly evenly across the globe [ 66 ]. This analysis also predicted that 37% of MDR isolates in this study had spread person-to-person, which is probably a vast underestimate of how frequently MDR is transmitted once evolved [ 66 ].
Geographic movement of people is also an important consideration with regard to person-to-person transmission. Further examination of the MDR clades from Manson et al. [ 66 ] revealed that they included widespread international, and even intercontinental, dissemination of strains that were separated by as few as four SNPs, probably due to spread via international travel [ 67 ]. Even within a single province in South Africa, Nelson et al. [ 68 ] showed, using genomic sequence data and global positioning system coordinates, that many cases of person-to-person transmission (with ≤ 5 SNPs) of XDR-TB occur between people living a median of 108 km apart, pointing to migration between urban and rural areas as a driver of TB spread. Collectively, these studies reinforce the idea that the geographic movement of people must be taken into consideration in any strategy for controlling the spread of TB resistance.
Ordering of the acquisition of resistance and compensatory mutations
Recent WGS studies have helped to illuminate the steps or ‘fitness landscape’ through which M. tuberculosis develops and compensates for drug resistance. Several studies [ 66 , 69 , 70 ] have shown that the order of acquisition of drug-resistance mutations in complex resistance cases is partly constrained in clinical M. tuberculosis . For example, in MDR-TB, isoniazid resistance (most often involving a katG S315T mutation) overwhelmingly evolves prior to resistance to rifampicin and second-line drugs. This was first shown using regional datasets from South Africa [ 69 ] and Argentina [ 70 ], and recently confirmed by Manson et al. [ 66 ] using a global dataset of 5310 strains. In the study by Manson et al. [ 66 ], this ordering was shown to hold true over 95% of the time, even for distinct global regions and time frames, including times when both rifampicin and isoniazid were in use, suggesting that the earlier introduction of isoniazid in the 1950s was not the major contributor to this effect. It was also shown that inhA promoter mutations that confer isoniazid resistance (such as those observed by Perdigão et al. [ 165 ] in Portugal) were acquired earlier than rifampicin mutations, although the number of samples harboring these mutations was much smaller. Further studies are necessary to determine whether isoniazid preventive monotherapy, which is one of the treatments for latent tuberculosis, may account for some of this effect, as this could result in a background level of increased isoniazid monoresistance.
Compensatory mutations that potentially ease fitness effects caused by resistance often occur after the evolution of primary resistance. This phenomenon was reviewed by Fonseca et al. [ 71 ], and examples include mutations in the ahpC promoter region and the rpoC / rpoA genes, which act as compensatory mutations for isoniazid and rifampicin resistance, respectively. Newer WGS work has pointed to several novel compensatory mutations in M. tuberculosis , particularly for rifampicin resistance. Comas et al. [ 72 ] identified a set of compensatory mutations in the rpoB gene that conferred high competitive fitness in vitro and were also found frequently in clinical populations. In a large-scale analysis of 1000 strains from Russia, Casali et al. [ 73 ] examined strains with primary resistance mutations in rpoB and identified accompanying compensatory mutations in rpoA and rpoC . Cohen et al. [ 69 ] identified putative rifampicin compensatory mutations that are present in South African strains by searching for rpoA , rpoB , and rpoC mutations that evolved only after or concurrent with rifampicin resistance-conferring mutations. A recent study of highly resistant M. tuberculosis strains from Central Asia confirmed that the presence of compensatory mutations, particularly those compensating for the fitness cost of mutations that confer rifampicin resistance, is associated with transmission success and higher drug-resistance rates [ 74 ]. Beyond rifampicin resistance compensation, Coll et al. [ 59 ] identified mutations in pncB2 that may compensate for pyrazinamide resistance conferred by pncA , and similarly, mutations in thyX-hsdS.1 (the thyX promoter) that may compensate for PAS resistance conferred by thyA ; however, experimental validation of these potential compensatory relationships is needed. Even fewer studies have identified stepping-stone mutations in M. tuberculosis , which emerge prior to higher-level resistance mutations. Cohen et al. [ 69 ] found that ubiA mutations emerge in a stepping-stone fashion prior to more classic embB mutations that confer ethambutol resistance. Safi et al. [ 15 ] also showed in vitro that multi-step selection involving ubiA , aftA , embB , and embC is required to achieve the highest levels of ethambutol resistance.
The challenge of mixed infections
Although WGS approaches have great sensitivity in detecting cases of recent transmission, reconstructing the details of transmission networks with WGS [ 166 , 167 , 168 ] can be difficult. Transmission network mapping is highly dependent on sampling density and studies rarely, if ever, comprehensively sample an outbreak or the extent of within-host diversity. It is also becoming clear, from the prevalence of very close relationships between isolates from patients who have no other direct epidemiological connections, that transmission may largely result from casual contact in community settings [ 169 ]. In addition, the phylogenetic reconstruction of transmission networks can be especially challenging, particularly because of the very close relationships between strains and the slow rate of evolution of M. tuberculosis [ 92 , 125 , 126 , 127 , 128 ].
Mixed infections represent a major challenge for understanding drug-resistance evolution within patients [ 153 , 158 , 159 ]. It can be straightforward to disambiguate co-infections of strains from different lineages, but mixed infections involving strains that have few genetic differences can also occur, making these strains difficult to distinguish. Köser et al. [ 75 ] used WGS for rapid drug-susceptibility testing of a patient with XDR-TB, and determined that the patient carried two different XDR-TB Beijing strains with differing resistance mutations. In a study by Liu et al. [ 76 ], three dominant sub-clones differing by 10–14 SNPs were detected within a single patient, each with different resistance patterns and probably different anatomical distributions. Also, co-infection by strains with differing resistance patterns may yield misleading composite views of resistance; for example, co-infection with two MDR-TB strains—one with quinolone resistance and the other with aminoglycoside resistance—may be mistaken for infection with a single XDR-TB strain.
Furthermore, newer data suggest that there can be genetic heterogeneity among M. tuberculosis isolates from different parts of the body, potentially leading to incomplete views of drug resistance within a patient (Fig. 1 ). In a study by Lieberman et al. [ 77 ], the authors observed evidence for both within-host evolution and mixed infection by piecing together the genetic variation observed among M. tuberculosis isolates from multiple post-mortem biopsies from the same patient. Another recent study by Dheda et al. [ 78 ] showed that drug concentrations at seven body sites were inversely correlated with the MIC of the bacteria isolated from these sites. Sequencing and comparison to pre-treatment and serial sputum samples suggested ongoing acquired resistance and differential evolution across sites [ 78 ]. These findings underscore the limitations of diagnosing or studying the evolution of drug-resistant M. tuberculosis using a single patient specimen. However, they also show the promise of WGS for informing interventions related to drug delivery, dosing, and diagnostics, thereby helping to prevent the development of acquired resistance within a patient. More research in this area is needed to determine the breadth and scope of mixed infections among patients with active TB, their contribution to changing drug-resistance patterns over time, and the role of spatial heterogeneity in the evolution of drug resistance.
Challenges to predicting drug resistance accurately from clinical specimens using current culture-dependent molecular diagnostics. The left panel depicts an expectorated sputum sample, which may not accurately represent the microbiologic diversity within the source patient. Culturing this sample (center panel) introduces further biases between faster- and slower-growing strains, such that faster-growing strains are over-represented within the cultured sample. Genomic DNA that is isolated and sequenced is input to computer algorithms that determine the genomic content, including the identification of drug-resistance mutations. However, disambiguating samples that contain mixed strains or detecting heteroresistance remains a computational challenge. The left panel was adapted from Ford et al. [ 170 ], with permission from Elsevier
From bench to bedside: promise and challenges
Given that the failure to identify and treat patients who have drug-resistant TB leads to increased mortality, spread of resistant strains, and gain of additional drug resistance [ 171 ], there is a critical need to diagnose resistant M. tuberculosis in patients rapidly. Several important molecular diagnostic platforms have been established for the identification of M. tuberculosis and drug resistance within this organism, but they are limited to the identification of a defined subset of resistance mutations [ 172 ], do not always include the earliest-arising mutations that precede MDR [ 66 ], and do not provide knowledge that is useful in determining whether a patient has been re-infected, whether the patient has a recurrent or mixed infection, or whether a particular infection represents a transmission event. WGS holds significant potential to modernize the TB laboratory and improve upon TB management [ 173 ], and this topic has been reviewed previously [ 173 , 174 ]. To date, WGS has been primarily applied as a clinical tool to achieve two goals: first, to detect M. tuberculosis within a clinical sample, and second, to detect resistance mutations and predict resistance patterns so that appropriate treatment can be provided. In order to provide clinically useful information, a diagnostic platform must be rapid. Historically, WGS has relied upon an input of pure mycobacterial cultures, which is time-consuming (requiring multiple weeks) and therefore of less clinical utility. Several investigations have attempted to address this issue by using earlier culture inputs or by attempting culture-independent, direct sequencing from clinical specimens [ 80 , 82 , 175 ]. In a rapid, yet still culture-dependent method, Pankhurst et al. [ 80 ] prospectively compared real-time WGS of “early positive liquid cultures” to routine M. tuberculosis diagnostics, and found that WGS achieved a faster time to diagnosis at a lower cost.
Although the advances achieved using WGS are promising, several hurdles must be overcome before it can be put into practical use in the clinic (Fig. 1 ). Requirements for costly equipment, technical expertise, and substantial computational resources present challenges to implementation [ 173 ]. Direct sequencing of patient samples has revealed that the vast majority of DNA present is from the patient or from non-mycobacterial prokaryotes, with variable quantities of mycobacterial DNA present. Doughty et al. [ 81 ] performed a pilot study demonstrating the feasibility of direct sequencing using a benchtop sequencer (Illumina MiSeq, San Diego, CA) and sputum samples from eight patients. Although they were able to identify the presence of M. tuberculosis , the low depth of sequencing coverage of the genome (0.002 to 0.7x) prevented drug-susceptibility prediction. Separately, Brown et al. [ 176 ] performed an enrichment step with biotinylated RNA baits prior to direct sequencing of sputum, resulting in higher quality data (> 20x depth and > 90% coverage) that allowed the identification of resistance mutations.
Using a targeted DNA enrichment strategy to study 43 individuals with active pulmonary TB, Doyle et al. [ 177 ] compared WGS directly from sputum with mycobacterial growth indicator tube (MGIT) WGS. Although direct sputum sequencing was able to identify drug resistance much more rapidly than MGIT WGS, only 74% of sputum samples yielded interpretable WGS data (vs 100% from MGIT); thus, additional optimization of these methods is needed to increase the sensitivity of this approach. Similarly, in a recent study, the use of pyrosequencing of a concentrated sputum sediment (rather than from sputum directly), dramatically shortened the time to initiation of an MDR-treatment regimen [ 178 ].
One promising technology that could change clinical WGS is long-read sequencing using the Oxford Nanopore Technologies (ONT; Oxford, UK) platform. An advantage of ONT is the ability to allow sequencing to continue until sufficient coverage of the genome has been obtained, potentially solving the problem of low or variable amounts of M. tuberculosis in clinical samples [ 82 ]. Early ONT studies have shown promise in identifying antimicrobial-resistance genes in different bacterial species [ 179 ]. Unfortunately, at present, both the high error rate of ONT MinION and potential difficulties with GC-rich regions limit the utility of this technology; thus, improvements in accuracy are necessary to enable the identification of resistance associated with point mutations [ 179 ]. ONT metagenomic sequencing has been successfully applied to improve pathogen detection and antimicrobial-resistance testing in other clinical settings [ 180 ]; however, to date, applications of this technology to M. tuberculosis have been limited to pre-clinical research [ 82 ].
Despite these challenges, WGS offers several advantages over the technologies that are currently employed for diagnosis and epidemiological monitoring of TB. Using WGS directly on patient sputum could reduce the turnaround time for diagnosis and determination of antibiotic-resistance status from weeks to hours [ 61 , 159 ], and would prevent the introduction of culture-induced biases. The depth of information provided by WGS could be used to identify whether an individual harbors multiple co-infecting strains [ 106 , 160 ] and to distinguish recurrent infection as either relapse or re-infection [ 174 , 181 ]. In addition, WGS could provide real-time epidemiological information that could be useful for understanding patterns of drug resistance and for establishing chains of transmission [ 174 ]. Encouragingly, the high levels of concordance observed between the genotypes and phenotypes of clinical samples indicate that WGS can provide high accuracy for both diagnosing TB and informing treatment options [ 113 ]. Finally, WGS of patient samples would provide a high level of convenience, by combining diagnosis, resistance profiling, and epidemiological analysis into a single test [ 85 ]. Given these advantages, the WHO has recently published a technical guide for the implementation of next-generation sequencing (NGS) technologies for the detection of drug resistance in M. tuberculosis [ 182 ].
Routine whole-genome sequencing of mycobacterial isolates
In 2017, England became the first nation to launch routine WGS of all prospectively identified M. tuberculosis clinical isolates [ 183 ]. Sponsored by Public Health England (PHE), prospective WGS is being performed on all positive mycobacterial cultures. Within 5–7 days of receipt of the culture from the reference lab, data will be provided on the mycobacterial species, the predicted drug susceptibility, and the molecular epidemiology of the strains. If, from the sequence analysis, a strain is predicted to be fully susceptible to first-line antitubercular drugs, phenotypic drug-susceptibility testing (DST) will no longer be performed routinely. However, if drug resistance to any first-line drug is identified, then phenotypic DST will follow. Beyond drug-susceptibility prediction, these efforts will have profound implications for TB control because WGS data can be used for real-time molecular epidemiology in this context.
Given the high sensitivity of WGS in detecting drug resistance to first-line TB drugs [ 60 ], similar algorithms utilizing WGS to predict susceptibility (rather than to identify drug resistance) for first-line drugs, in lieu of phenotypic DST, have been endorsed in the Netherlands and in New York [ 60 ]. It seems highly likely that these kinds of efforts would be helpful in higher-burden TB settings than those mentioned here, but the feasibility of this approach has not yet been established, from either a practical or economic standpoint, in settings where the numbers of drug-resistant TB cases are high.
Conclusions and future directions
Since the first applications of WGS to M. tuberculosis in 1998, WGS techniques have greatly accelerated our understanding of drug-resistance mechanisms in this pathogen. Importantly, WGS studies now indicate that, for many drugs, the vast majority of resistance is explained by known mutations. The increasing availability of whole-genome sequences from phenotypically diverse M. tuberculosis , combined with improved GWAS algorithms, is enabling the discovery of the remaining determinants of unexplained resistance. In addition, WGS has provided valuable insight into how resistance mutations evolve and spread. It is clear that both de novo acquisition of resistance mutations and clonal transmission are critical factors in the spread of drug-resistant TB.
Furthermore, WGS investigations have revealed that there is a specific order in which drug-resistance mutations are acquired: isoniazid resistance is almost always acquired before rifampicin resistance, which has significant implications for the design of diagnostic tests. Within individual patients, WGS studies have highlighted that mixed infections are common, and often represent important intermediates in the evolution of drug resistance.
WGS also holds great promise for revolutionizing the rapid clinical diagnosis of TB in the future. Although there are still substantial technical hurdles, WGS can be used to diagnose the presence of M. tuberculosis rapidly, as well as to pinpoint appropriate antibiotic treatment regimens by identifying the complement of M. tuberculosis drug-resistance mutations that are present within a clinical sample. Indeed, improvements in the prediction of drug susceptibility with WGS may obviate the need for phenotypic culture methods, especially for first-line drugs.
Although WGS offers many benefits, targeted NGS, in which sequence data are obtained from only a focused panel of genes or genetic regions rather than from the entire genome, is gaining momentum [ 184 ]. One of the advantages of targeted NGS over WGS is that it can be performed directly on clinical specimens and is, therefore, faster than culture-based WGS. Other advantages include reduction in both labor and computational efforts and reduced costs. The potential offered by the application of targeted NGS to the prediction of drug resistance from genomic data is self-evident. Nevertheless, it seems that WGS would have greater discriminatory power than targeted NGS for molecular epidemiology purposes.
Ultimately, the use of WGS is expected to continue to advance our understanding of M. tuberculosis drug resistance. Furthermore, its practical use in clinical settings holds great potential to improve public health through real-time molecular epidemiology tracking, to identify global hotspots of drug-resistance emergence, and to facilitate the development of improved approaches for the diagnosis and treatment of drug-resistant TB.
Abbreviations
Drug-susceptibility testing
Genome-wide association study
Multidrug-resistant
Minimum inhibitory concentration
Next generation sequencing
Oxford Nanopore Technologies
Para-aminosalicylic acid
Rifampicin-resistant-TB
Single nucleotide polymorphism
Tuberculosis
Whole-genome sequencing
World Health Organization (WHO)
Extensively drug-resistant
World Health Organization. Global Tuberculosis Report 2018. Geneva: WHO; 2018. http://apps.who.int/iris/bitstream/handle/10665/274453/9789241565646-eng.pdf?ua=1 . Accessed 9 July 2019
Google Scholar
Streptomycin in Tuberculosis Trials Committee. Streptomycin treatment for pulmonary tuberculosis. Br Med J. 1948;2:769–82 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2091872/pdf/brmedj03701-0007.pdf . Accessed 9 July 2019.
Article Google Scholar
World Health Organization. Global Tuberculosis Report 2016. Geneva: WHO; 2016. https://apps.who.int/iris/bitstream/handle/10665/250441/9789241565394-eng.pdf?sequence=1 . Accessed 9 July 2019
World Health Organization. Global Tuberculosis Report 2017. Geneva: WHO; 2017. https://www.who.int/tb/publications/global_report/gtbr2017_main_text.pdf . Accessed 9 July 2019
World Health Organization. WHO treatment guidelines for drug-resistant tuberculosis. 2016 update. Geneva: WHO; 2016. https://apps.who.int/iris/bitstream/handle/10665/250125/9789241549639-eng.pdf?sequence=1 . Accessed 9 July 2019
Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston MJ, et al. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis . Lancet. 1993;341:647–50.
Article CAS PubMed Google Scholar
Farhat MR, Shapiro BJ, Kieser KJ, Sultana R, Jacobson KR, Victor TC, et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis . Nat Genet. 2013;45:1183–9.
Article CAS PubMed PubMed Central Google Scholar
Heym B, Alzari PM, Honoré N, Cole ST. Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis . Mol Microbiol. 1995;15:235–45.
Rozwarski DA, Grant GA, Barton DH, Jacobs WR, Sacchettini JC. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis . Science. 1998;279:98–102.
Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, et al. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis . Science. 1994;263:227–30.
Sreevatsan S, Pan X, Zhang Y, Kreiswirth BN, Musser JM. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob Agents Chemother. 1997;41:636–40.
Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, et al. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis . Antimicrob Agents Chemother. 1997;41:540–3.
Zhang S, Chen J, Shi W, Liu W, Zhang W, Zhang Y. Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis . Emerg Microbes Infect. 2013;2:e34.
Shi W, Zhang X, Jiang X, Yuan H, Lee JS, Barry CE, et al. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis . Science. 2011;333:1630–2.
Safi H, Lingaraju S, Amin A, Kim S, Jones M, Holmes M, et al. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-D-arabinose biosynthetic and utilization pathway genes. Nat Genet. 2013;45:1190–7.
Alcaide F, Pfyffer GE, Telenti A. Role of embB in natural and acquired resistance to ethambutol in mycobacteria. Antimicrob Agents Chemother. 1997;41:2270–3.
Maruri F, Sterling TR, Kaiga AW, Blackman A, van der Heijden YF, Mayer C, et al. A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J Antimicrob Chemother. 2012;67:819–31.
Takiff HE, Salazar L, Guerrero C, Philipp W, Huang WM, Kreiswirth B, et al. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother. 1994;38:773–80.
Andries K, Verhasselt P, Guillemont J, Göhlmann HWH, Neefs J-M, Winkler H, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis . Science. 2005;307:223–7.
Almeida D, Ioerger T, Tyagi S, Li SY, Mdluli K, Andries K, et al. Mutations in pepQ confer low-level resistance to bedaquiline and clofazimine in Mycobacterium tuberculosis . Antimicrob Agents Chemother. 2016;60:4590–9.
Hartkoorn RC, Uplekar S, Cole ST. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis . Antimicrob Agents Chemother. 2014;58:2979–81.
Article PubMed PubMed Central CAS Google Scholar
Andries K, Villellas C, Coeck N, Thys K, Gevers T, Vranckx L, et al. Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS One. 2014. https://doi.org/10.1371/journal.pone.0102135 .
Hillemann D, Rüsch-Gerdes S, Richter E. In vitro-selected linezolid-resistant Mycobacterium tuberculosis mutants. Antimicrob Agents Chemother. 2008;52:800–1.
Beckert P, Hillemann D, Kohl TA, Kalinowski J, Richter E, Niemann S, et al. rplC T460C identified as a dominant mutation in linezolid-resistant Mycobacterium tuberculosis strains. Antimicrob Agents Chemother. 2012;56:2743–5.
Desjardins CA, Cohen KA, Munsamy V, Abeel T, Maharaj K, Walker BJ, et al. Genomic and functional analyses of Mycobacterium tuberculosis strains implicate ald in D-cycloserine resistance. Nat Genet. 2016;48:544–51.
Cáceres NE, Harris NB, Wellehan JF, Feng Z, Kapur V, Barletta RG. Overexpression of the D-alanine racemase gene confers resistance to D-cycloserine in Mycobacterium smegmatis . J Bacteriol. 1997;179:5046–55.
Article PubMed PubMed Central Google Scholar
Feng Z, Barletta RG. Roles of Mycobacterium smegmatis D-alanine:D-alanine ligase and D-alanine racemase in the mechanisms of action of and resistance to the peptidoglycan inhibitor D-cycloserine. Antimicrob Agents Chemother. 2003;47:283–91.
Chen JM, Uplekar S, Gordon SV, Cole ST. A point mutation in cycA partially contributes to the D-cycloserine resistance trait of Mycobacterium bovis BCG vaccine strains. PLoS One. 2012. https://doi.org/10.1371/journal.pone.0043467 .
Haver HL, Chua A, Ghode P, Lakshminarayana SB, Singhal A, Mathema B, et al. Mutations in genes for the F420 biosynthetic pathway and a nitroreductase enzyme are the primary resistance determinants in spontaneous in vitro-selected PA-824-resistant mutants of Mycobacterium tuberculosis . Antimicrob Agents Chemother. 2015;59:5316–23.
Kumar P, Kaushik A, Bell D, Chauhan V, Xia F, Stevens RL, et al. Mutation in an unannotated protein confers carbapenem resistance in Mycobacterium tuberculosis . Antimicrob Agents Chemother. 2017. https://doi.org/10.1128/AAC.02234-16 .
Suzuki Y, Katsukawa C, Tamaru A, Abe C, Makino M, Mizuguchi Y, et al. Detection of kanamycin-resistant Mycobacterium tuberculosis by identifying mutations in the 16S rRNA gene. J Clin Microbiol. 1998;36:1220–5.
CAS PubMed PubMed Central Google Scholar
Finken M, Kirschner P, Meier A, Wrede A, Böttger EC. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis : alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol Microbiol. 1993;9:1239–46.
Nair J, Rouse DA, Bai GH, Morris SL. The rpsL gene and streptomycin resistance in single and multiple drug-resistant strains of Mycobacterium tuberculosis . Mol Microbiol. 1993;10:521–7.
Honoré N, Cole ST. Streptomycin resistance in mycobacteria. Antimicrob Agents Chemother. 1994;38:238–42.
Meier A, Kirschner P, Bange FC, Vogel U, Böttger EC. Genetic alterations in streptomycin-resistant Mycobacterium tuberculosis : mapping of mutations conferring resistance. Antimicrob Agents Chemother. 1994;38:228–33.
Douglass J, Steyn LM. A ribosomal gene mutation in streptomycin-resistant Mycobacterium tuberculosis isolates. J Infect Dis. 1993;167:1505–6.
Wong SY, Lee JS, Kwak HK, Via LE, Boshoff HIM, Barry CE. Mutations in gidB confer low-level streptomycin resistance in Mycobacterium tuberculosis . Antimicrob Agents Chemother. 2011;55:2515–22.
Morlock GP, Metchock B, Sikes D, Crawford JT, Cooksey RC. ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2003;47:3799–805.
Baulard AR, Betts JC, Engohang-Ndong J, Quan S, McAdam RA, Brennan PJ, et al. Activation of the pro-drug ethionamide is regulated in mycobacteria. J Biol Chem. 2000;275:28326–31.
CAS PubMed Google Scholar
Zheng J, Rubin EJ, Bifani P, Mathys V, Lim V, Au M, et al. Para-aminosalicylic acid is a prodrug targeting dihydrofolate reductase in Mycobacterium tuberculosis . J Biol Chem. 2013;288:23447–56.
Mathys V, Wintjens R, Lefevre P, Bertout J, Singhal A, Kiass M, et al. Molecular genetics of para-aminosalicylic acid resistance in clinical isolates and spontaneous mutants of Mycobacterium tuberculosis . Antimicrob Agents Chemother. 2009;53:2100–9.
Rengarajan J, Sassetti CM, Naroditskaya V, Sloutsky A, Bloom BR, Rubin EJ. The folate pathway is a target for resistance to the drug para-aminosalicylic acid (PAS) in mycobacteria. Mol Microbiol. 2004;53:275–82.
Fivian-Hughes AS, Houghton J, Davis EO, Hill M, Nw L. Mycobacterium tuberculosis thymidylate synthase gene thyX is essential and potentially bifunctional, while thyA deletion confers resistance to p-aminosalicylic acid. Microbiology. 2012;158:308–18.
Zhang X, Liu L, Zhang Y, Dai G, Huang H, Jin Q. Genetic determinants involved in p-aminosalicylic acid resistance in clinical isolates from tuberculosis patients in northern China from 2006 to 2012. Antimicrob Agents Chemother. 2015;59:1320–4.
Zaunbrecher MA, Sikes RD, Metchock B, Shinnick TM, Posey JE. Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis . Proc Natl Acad Sci U S A. 2009;106:20004–9.
Maus CE, Plikaytis BB, Shinnick TM. Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis . Antimicrob Agents Chemother. 2005;49:571–7.
World Health Organization. Rapid Communication: key changes to treatment of multidrug- and rifampicin-resistant tuberculosis (MDR/RR-TB). Geneva: WHO; 2018. https://www.who.int/tb/publications/2018/WHO_RapidCommunicationMDRTB.pdf?ua=1 . Accessed 9 July 2019
Diacon AH, Donald PR, Pym A, Grobusch M, Patientia RF, Mahanyele R, et al. Randomized pilot trial of eight weeks of bedaquiline (TMC207) treatment for multidrug-resistant tuberculosis: long-term outcome, tolerability, and effect on emergence of drug resistance. Antimicrob Agents Chemother. 2012;56:3271–6.
Diacon AH, Pym A, Grobusch MP, de Los Rios JM, Gotuzzo E, Vasilyeva I, et al. Multidrug-resistant tuberculosis and culture conversion with bedaquiline. N Engl J Med. 2014;371:723–32.
Article PubMed CAS Google Scholar
Matsumoto M, Hashizume H, Tomishige T, Kawasaki M, Tsubouchi H, Sasaki H, et al. OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med. 2006;3:2131–44.
Article CAS Google Scholar
Gler MT, Skripconoka V, Sanchez-Garavito E, Xiao H, Cabrera-Rivero JL, Vargas-Vasquez DE, et al. Delamanid for multidrug-resistant pulmonary tuberculosis. N Engl J Med. 2012;366:2151–60.
Bloom BR, Atun R, Cohen T, Dye C, Fraser H, Gomez GB, et al. Tuberculosis. In: Holmes KK, Bertozzi S, Bloom BR, et al., editors. Major infectious diseases. Washington DC: The International Bank for Reconstruction and Development/The World Bank; 2017. https://www.ncbi.nlm.nih.gov/books/NBK525174/ . Accessed 9 July 2019.
Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44.
Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, Peterson J, et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol. 2002;184:5479–90.
Young DB. Blueprint for the white plague. Nature. 1998;393:515–6.
Cabibbe AM, Walker TM, Niemann S, Cirillo DM. Whole genome sequencing of Mycobacterium tuberculosis . Eur Respir J. 2018;52:537–45.
Zhang H, Li D, Zhao L, Fleming J, Lin N, Wang T, et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat Genet. 2013;45:1255–60.
Walker TM, Kohl TA, Omar SV, Hedge J, Del Ojo EC, Bradley P, et al. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis. 2015;3099:1–10.
Coll F, Phelan J, Hill-Cawthorne GA, Nair MB, Mallard K, Ali S, et al. Genome-wide analysis of multi- and extensively drug-resistant Mycobacterium tuberculosis . Nat Genet. 2018;50:307–16.
Article PubMed Google Scholar
The CRyPTIC Consortium and the 100,000 Genomes Project. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N Engl J Med. 2018;379:1403–15.
Eldholm V, Norheim G, von der Lippe B, Kinander W, Dahle UR, Caugant DA, et al. Evolution of extensively drug-resistant Mycobacterium tuberculosis from a susceptible ancestor in a single patient. Genome Biol. 2014. https://doi.org/10.1186/s13059-014-0490-3 .
Trauner A, Liu Q, Via LE, Liu X, Ruan X, Liang L, et al. The within-host population dynamics of Mycobacterium tuberculosis vary with treatment efficacy. Genome Biol. 2017. https://doi.org/10.1186/s13059-017-1196-0 .
Nikolayevskyy V, Kranzer K, Niemann S, Drobniewski F. Whole genome sequencing of Mycobacterium tuberculosis for detection of recent transmission and tracing outbreaks: a systematic review. Tuberculosis. (Edinb.). 2016;98:77–85.
Ioerger TR, Feng Y, Chen X, Dobos KM, Victor TC, Streicher EM, et al. The non-clonality of drug resistance in Beijing-genotype isolates of Mycobacterium tuberculosis from the Western Cape of South Africa. BMC Genomics. 2010. https://doi.org/10.1186/1471-2164-11-670 .
Shah NS, Auld SC, Brust JCM, Mathema B, Ismail N, Moodley P, et al. Transmission of extensively drug-resistant tuberculosis in South Africa. N Engl J Med. 2017;376:243–53.
Manson AL, Cohen KA, Abeel T, Desjardins CA, Armstrong DT, Barry CE, et al. Genomic analysis of globally diverse Mycobacterium tuberculosis strains provides insights into the emergence and spread of multidrug resistance. Nat Genet. 2017;49:395–402.
Cohen KA, Manson AL, Abeel T, Desjardins CA, Chapman SB, Hoffner S, et al. Extensive global movement of multidrug-resistant M. tuberculosis strains revealed by whole-genome analysis. Thorax. 2019. https://doi.org/10.1136/thoraxjnl-2018-211616 .
Nelson KN, Shah NS, Mathema B, Ismail N, Brust JCM, Brown TS, et al. Spatial patterns of extensively drug-resistant tuberculosis transmission in KwaZulu-Natal. South Africa. J Infect Dis. 2018;218:1964–73.
PubMed Google Scholar
Cohen KA, Abeel T, Manson McGuire A, Desjardins CA, Munsamy V, Shea TP, et al. Evolution of extensively drug-resistant tuberculosis over four decades: whole genome sequencing and dating analysis of Mycobacterium tuberculosis isolates from KwaZulu-Natal. PLoS Med. 2015. https://doi.org/10.1371/journal.pmed.1001880 .
Eldholm V, Monteserin J, Rieux A, Lopez B, Sobkowiak B, Ritacco V, et al. Four decades of transmission of a multidrug-resistant Mycobacterium tuberculosis outbreak strain. Nat Commun. 2015. https://doi.org/10.1038/ncomms8119 .
Fonseca JD, Knight GM, McHugh TD. The complex evolution of antibiotic resistance in Mycobacterium tuberculosis . Int J Infect Dis. 2015;32:94–100.
Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2012;44:106–10.
Casali N, Nikolayevskyy V, Balabanova Y, Harris SR, Ignatyeva O, Kontsevaya I, et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 2014;46:279–86.
Merker M, Barbier M, Cox H, Rasigade J-P, Feuerriegel S, Kohl TA, et al. Compensatory evolution drives multidrug-resistant tuberculosis in Central Asia. Elife. 2018. https://doi.org/10.7554/eLife.38200 .
Köser CU, Bryant JM, Becq J, Török ME, Ellington MJ, Marti-Renom MA, et al. Whole-genome sequencing for rapid susceptibility testing of M. tuberculosis . N Engl J Med. 2013;369:290–2.
Liu Q, Via LE, Luo T, Liang L, Liu X, Wu S, et al. Within patient microevolution of Mycobacterium tuberculosis correlates with heterogeneous responses to treatment. Sci Rep. 2015. https://doi.org/10.1038/srep17507 .
Lieberman TD, Wilson D, Misra R, Xiong LL, Moodley P, Cohen T, et al. Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated Mycobacterium tuberculosis . Nat Med. 2016;22:1470–4.
Dheda K, Lenders L, Magombedze G, Srivastava S, Raj P, Arning E, et al. Drug-penetration gradients associated with acquired drug resistance in patients with tuberculosis. Am J Respir Crit Care Med. 2018;198:1208–19.
Sobkowiak B, Glynn JR, Houben RMGJ, Mallard K, Phelan JE, Guerra-Assunção JA, et al. Identifying mixed Mycobacterium tuberculosis infections from whole genome sequence data. BMC Genomics. 2018. https://doi.org/10.1186/s12864-018-4988-z .
Pankhurst LJ, Del Ojo EC, Votintseva AA, Walker TM, Cole K, Davies J, et al. Rapid, comprehensive, and affordable mycobacterial diagnosis with whole-genome sequencing: a prospective study. Lancet Respir Med. 2016;4:49–58.
Doughty EL, Sergeant MJ, Adetifa I, Antonio M, Pallen MJ. Culture-independent detection and characterisation of Mycobacterium tuberculosis and M. africanum in sputum samples using shotgun metagenomics on a benchtop sequencer. PeerJ. 2014. doi: https://doi.org/10.7717/peerj.585 .
Votintseva AA, Bradley P, Pankhurst L, Del Ojo EC, Loose M, Nilgiriwala K, et al. Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J Clin Microbiol. 2017;55:1285–98.
Liu X, Gutacker MM, Musser JM, Fu Y-X. Evidence for recombination in Mycobacterium tuberculosis . J Bacteriol. 2006;188:8169–77.
Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis : 1998 update. Tuber Lung Dis. 1998;79:3–29.
Simons SO, van Ingen J, van der Laan T, Mulder A, Dekhuijzen PNR, Boeree MJ, et al. Validation of pncA gene sequencing in combination with the mycobacterial growth indicator tube method to test susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Clin Microbiol. 2012;50:428–34.
Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol. 2010;48:229–37.
Chen J, Zhang S, Cui P, Shi W, Zhang W, Zhang Y. Identification of novel mutations associated with cycloserine resistance in Mycobacterium tuberculosis . J Antimicrob Chemother. 2017;72:3272–6.
Zampieri M, Szappanos B, Buchieri MV, Trauner A, Piazza I, Picotti P, et al. High-throughput metabolomic analysis predicts mode of action of uncharacterized antimicrobial compounds. Sci Transl Med. 2018. https://doi.org/10.1126/scitranslmed.aal3973 .
Gagneux S. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis . Science. 2006;312:1944–6.
Bergval IL, Schuitema ARJ, Klatser PR, Anthony RM. Resistant mutants of Mycobacterium tuberculosis selected in vitro do not reflect the in vivo mechanism of isoniazid resistance. J Antimicrob Chemother. 2009;64:515–23.
Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, Cohen T, et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet. 2013;45:784–90.
Ford CB, Lin PL, Chase MR, Shah RR, Iartchouk O, Galagan J, et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet. 2011;43:482–6.
Papaventsis D, Casali N, Kontsevaya I, Drobniewski F, Cirillo DM, Nikolayevskyy V. Whole genome sequencing of Mycobacterium tuberculosis for detection of drug resistance: a systematic review. Clin Microbiol Infect. 2017;23:61–8.
Gygli SM, Keller PM, Ballif M, Blöchliger N, Hömke R, Reinhard M, et al. Whole genome sequencing for drug resistance profile prediction in Mycobacterium tuberculosis . Antimicrob Agents Chemother. 2019. https://doi.org/10.1128/AAC.02175-18 .
Cohen KA, El-Hay T, Wyres KL, Weissbrod O, Munsamy V, Yanover C, et al. Paradoxical hypersusceptibility of drug-resistant Mycobacterium tuberculosis to β-lactam antibiotics. EBioMedicine. 2016;9:170–9.
World Health Organization. Companion handbook to the WHO guidelines for the programmatic management of drug-resistant tuberculosis. Geneva: WHO; 2014. https://apps.who.int/iris/bitstream/handle/10665/130918/9789241548809_eng.pdf?sequence=1 . Accessed 9 July 2019
Power RA, Parkhill J, de Oliveira T. Microbial genome-wide association studies: lessons from human GWAS. Nat Rev Genet. 2017;18:41–50.
Farhat MR, Freschi L, Calderon R, Ioerger T, Snyder M, Meehan CJ, et al. Genome wide association with quantitative resistance phenotypes in Mycobacterium tuberculosis reveals novel resistance genes and regulatory regions. Nat Commun. 2019. https://doi.org/10.1038/s41467-019-10110-6 .
Earle SG, Wu C-H, Charlesworth J, Stoesser N, Gordon NC, Walker TM, et al. Identifying lineage effects when controlling for population structure improves power in bacterial association studies. Nat Microbiol. 2016. https://doi.org/10.1038/nmicrobiol.2016.41 .
Jaillard M, Lima L, Tournoud M, Mahé P, van Belkum A, Lacroix V, et al. A fast and agnostic method for bacterial genome-wide association studies: bridging the gap between k-mers and genetic events. PLoS Genet. 2018. https://doi.org/10.1371/journal.pgen.1007758 .
Lees JA, Vehkala M, Välimäki N, Harris SR, Chewapreecha C, Croucher NJ, et al. Sequence element enrichment analysis to determine the genetic basis of bacterial phenotypes. Nat Commun. 2016. https://doi.org/10.1038/ncomms12797 .
Alam MT, Petit RA, Crispell EK, Thornton TA, Conneely KN, Jiang Y, et al. Dissecting vancomycin-intermediate resistance in staphylococcus aureus using genome-wide association. Genome Biol Evol. 2014;6:1174–85.
Collins C, Didelot X. A phylogenetic method to perform genome-wide association studies in microbes that accounts for population structure and recombination. PLoS Comput Biol. 2018. https://doi.org/10.1371/journal.pcbi.1005958 .
Barker D, Meade A, Pagel M. Constrained models of evolution lead to improved prediction of functional linkage from correlated gain and loss of genes. Bioinformatics. 2007;23:14–20.
Hicks ND, Yang J, Zhang X, Zhao B, Grad YH, Liu L, et al. Clinically prevalent mutations in Mycobacterium tuberculosis alter propionate metabolism and mediate multidrug tolerance. Nat Microbiol. 2018;3:1032–42.
Merker M, Kohl TA, Roetzer A, Truebe L, Richter E, Rüsch-Gerdes S, et al. Whole genome sequencing reveals complex evolution patterns of multidrug-resistant Mycobacterium tuberculosis Beijing strains in patients. PLoS One. 2013. https://doi.org/10.1371/journal.pone.0082551 .
Stucki D, Gagneux S. Single nucleotide polymorphisms in Mycobacterium tuberculosis and the need for a curated database. Tuberculosis (Edinb). 2013;93:30–9.
Reddy TBK, Riley R, Wymore F, Montgomery P, DeCaprio D, Engels R, et al. TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 2009;37:D499–508.
Gillespie JJ, Wattam AR, Cammer SA, Gabbard JL, Shukla MP, Dalay O, et al. PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infect Immun. 2011;79:4286–98.
Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. Tuberculosis drug resistance mutation database. PLoS Med. 2009. https://doi.org/10.1371/journal.pmed.1000002 .
Article PubMed Central CAS Google Scholar
Iwai H, Kato-Miyazawa M, Kirikae T, Miyoshi-Akiyama T. CASTB (the comprehensive analysis server for the Mycobacterium tuberculosis complex): a publicly accessible web server for epidemiological analyses, drug-resistance prediction and phylogenetic comparison of clinical isolates. Tuberculosis (Edinb). 2015;95:843–4.
Steiner A, Stucki D, Coscolla M, Borrell S, Gagneux S. KvarQ. Targeted and direct variant calling from fastq reads of bacterial genomes. BMC Genomics. 2014. doi: https://doi.org/10.1186/1471-2164-15-881 .
Bradley P, Gordon NC, Walker TM, Dunn L, Heys S, Huang B, et al. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis . Nat Commun. 2015. https://doi.org/10.1038/ncomms10063 .
Feuerriegel S, Schleusener V, Beckert P, Kohl TA, Miotto P, Cirillo DM, et al. PhyResSE: a web tool delineating Mycobacterium tuberculosis antibiotic resistance and lineage from whole-genome sequencing data. J Clin Microbiol. 2015;53:1908–14.
Coll F, McNerney R, Preston MD, Guerra-Assunção JA, Warry A, Hill-Cawthorne G, et al. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med. 2015. https://doi.org/10.1186/s13073-015-0164-0 .
Sekizuka T, Yamashita A, Murase Y, Iwamoto T, Mitarai S, Kato S, et al. TGS-TB: total genotyping solution for Mycobacterium tuberculosis using short-read whole-genome sequencing. PLoS One. 2015. https://doi.org/10.1371/journal.pone.0142951 .
Schleusener V, Köser CU, Beckert P, Niemann S, Feuerriegel S. Mycobacterium tuberculosis resistance prediction and lineage classification from genome sequencing: comparison of automated analysis tools. Sci Rep. 2017. https://doi.org/10.1038/srep46327 .
van Beek J, Haanperä M, Smit PW, Mentula S, Soini H. Evaluation of whole genome sequencing and software tools for drug susceptibility testing of Mycobacterium tuberculosis . Clin Microbiol Infect. 2019;25:82–6.
Ngo T-M, Teo Y-Y. Genomic prediction of tuberculosis drug-resistance: benchmarking existing databases and prediction algorithms. BMC Bioinformatics. 2019. https://doi.org/10.1186/s12859-019-2658-z .
ReSeqTB. Relational sequencing TB data platform. https://platform.reseqtb.org . Accessed 9 July 2019.
Starks AM, Avilés E, Cirillo DM, Denkinger CM, Dolinger DL, Emerson C, et al. Collaborative effort for a centralized worldwide tuberculosis relational sequencing data platform. Clin Infect Dis. 2015;61:S141–6.
Article PubMed Central Google Scholar
CRyPTIC. Comprehensive Resistance Prediction for Tuberculosis: an International Consortium. http://www.crypticproject.org . Accessed 9 July 2019.
Miotto P, Tessema B, Tagliani E, Chindelevitch L, Starks AM, Emerson C, et al. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis . Eur Respir J. 2017. https://doi.org/10.1183/13993003.01354-2017 .
Falush D, Bowden R. Genome-wide association mapping in bacteria? Trends Microbiol. 2006;14:353–5.
Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–46.
Guerra-Assunção J, Crampin A, Houben R, Mzembe T, Mallard K, Coll F, et al. Large scale population-based whole genome sequencing of Mycobacterium tuberculosis provides insights into transmission in a high prevalence area. Elife. 2015. https://doi.org/10.7554/eLife.05166 .
Bryant JM, Schürch AC, van Deutekom H, Harris SR, de Beer JL, de Jager V, et al. Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect Dis. 2013. https://doi.org/10.1186/1471-2334-13-110 .
Roetzer A, Diel R, Kohl TA, Rückert C, Nübel U, Blom J, et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med. 2013. https://doi.org/10.1371/journal.pmed.1001387 .
World Health Organization. Policy guidance on drug-susceptibility testing (DST) of second-line antituberculosis drugs. Geneva: WHO; 2008. https://apps.who.int/iris/bitstream/handle/10665/70500/WHO_HTM_TB_2008.392_eng.pdf?sequence=1 . Accessed 9 July 2019
Colangeli R, Jedrey H, Kim S, Connell R, Ma S, Chippada Venkata UD, et al. Bacterial factors that predict relapse after tuberculosis therapy. N Engl J Med. 2018;379:823–33.
Rinder H. Hetero-resistance: an under-recognised confounder in diagnosis and therapy? J Med Microbiol. 2001;50:1018–20.
Rinder H, Mieskes KT, Löscher T. Heteroresistance in Mycobacterium tuberculosis . Int J Tuberc Lung Dis. 2001;5:339–45.
Folkvardsen DB, Thomsen VØ, Rigouts L, Rasmussen EM, Bang D, Bernaerts G, et al. Rifampin heteroresistance in Mycobacterium tuberculosis cultures as detected by phenotypic and genotypic drug susceptibility test methods. J Clin Microbiol. 2013;51:4220–2.
Liang B, Tan Y, Li Z, Tian X, Du C, Li H, et al. Highly sensitive detection of isoniazid heteroresistance in Mycobacterium tuberculosis by DeepMelt assay. J Clin Microbiol. 2018. https://doi.org/10.1128/JCM.01239-17 .
Zhang D, Gomez JE, Chien J-Y, Haseley N, Desjardins CA, Earl AM, et al. Genomic analysis of the evolution of fluoroquinolone resistance in Mycobacterium tuberculosis prior to tuberculosis diagnosis. Antimicrob Agents Chemother. 2016;60:6600–8.
Phelan JE, Coll F, Bergval I, Anthony RM, Warren R, Sampson SL, et al. Recombination in pe/ppe genes contributes to genetic variation in Mycobacterium tuberculosis lineages. BMC Genomics. 2016. https://doi.org/10.1186/s12864-016-2467-y .
Akhter Y, Ehebauer MT, Mukhopadhyay S, Hasnain SE. The PE/PPE multigene family codes for virulence factors and is a possible source of mycobacterial antigenic variation: Perhaps more? Biochimie. 2012;94:110–6.
Farhat MR, Shapiro B, Sheppard SK, Colijn C, Murray M. A phylogeny-based sampling strategy and power calculator informs genome-wide associations study design for microbial pathogens. Genome Med. 2014. https://doi.org/10.1186/s13073-014-0101-7 .
Casali N, Broda A, Harris SR, Parkhill J, Brown T, Drobniewski F. Whole genome sequence analysis of a large isoniazid-resistant tuberculosis outbreak in London: a retrospective observational study. PLoS Med. 2016. https://doi.org/10.1371/journal.pmed.1002137 .
van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–9.
PubMed PubMed Central Google Scholar
Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–14.
Supply P, Magdalena J, Himpens S, Locht C. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol Microbiol. 1997;26:991–1003.
Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis . J Clin Microbiol. 2006;44:4498–510.
Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 2010;38:W326–31.
Oelemann MC, Diel R, Vatin V, Haas W, Rüsch-Gerdes S, Locht C, et al. Assessment of an optimized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J Clin Microbiol. 2007;45:691–7.
Chihota VN, Müller B, Mlambo CK, Pillay M, Tait M, Streicher EM, et al. Population structure of multi- and extensively drug-resistant Mycobacterium tuberculosis strains in South Africa. J Clin Microbiol. 2012;50:995–1002.
Gandhi NR, Brust JCM, Moodley P, Weissman D, Heo M, Ning Y, et al. Minimal diversity of drug-resistant Mycobacterium tuberculosis strains. South Africa. Emerg Infect Dis. 2014;20:394–401.
Bifani PJ, Plikaytis BB, Kapur V, Stockbauer K, Pan X, Lutfey ML, et al. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA. 1996;275:452–7.
Lutfey M, Della-Latta P, Kapur V, Palumbo LA, Gurner D, Stotzky G, et al. Independent origin of mono-rifampin-resistant Mycobacterium tuberculosis in patients with AIDS. Am J Respir Crit Care Med. 1996;153:837–40.
Post FA, Willcox PA, Mathema B, Steyn LM, Shean K, Ramaswamy SV, et al. Genetic polymorphism in Mycobacterium tuberculosis isolates from patients with chronic multidrug-resistant tuberculosis. J Infect Dis. 2004;190:99–106.
Roof I, Jajou R, Kamst M, Mulder A, de Neeling A, van Hunen R, et al. Prevalence and characterization of heterogeneous variable-number tandem-repeat clusters comprising drug-susceptible and/or variable resistant Mycobacterium tuberculosis complex isolates in the Netherlands from 2004 to 2016. J Clin Microbiol. 2018. https://doi.org/10.1128/JCM.00887-18 .
Witney AA, Bateson AL, Jindani A, Phillips PP, Coleman D, Stoker NG, et al. Use of whole-genome sequencing to distinguish relapse from reinfection in a completed tuberculosis clinical trial. BMC Med. 2017. https://doi.org/10.1186/s12916-017-0834-4 .
Van Rie A, Victor TC, Richardson M, Johnson R, Van Der Spuy GD, Murray EJ, et al. Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am J Respir Crit Care Med. 2005;172:636–42.
Warren RM, Victor TC, Streicher EM, Richardson M, Beyers N, Gey van Pittius NC, et al. Patients with active tuberculosis often have different strains in the same sputum specimen. Am J Respir Crit Care Med. 2004;169:610–4.
Hatherell H-A, Colijn C, Stagg HR, Jackson C, Winter JR, Abubakar I. Interpreting whole genome sequencing for investigating tuberculosis transmission: a systematic review. BMC Med. 2016. https://doi.org/10.1186/s12916-016-0566-x .
Sun G, Luo T, Yang C, Dong X, Li J, Zhu Y, et al. Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J Infect Dis. 2012;206:1724–33.
Mariam SH, Werngren J, Aronsson J, Hoffner S, Andersson DI. Dynamics of antibiotic resistant mycobacterium tuberculosis during long-term infection and antibiotic treatment. PLoS One. 2011. https://doi.org/10.1371/journal.pone.0021147 .
Manson AL, Abeel T, Galagan JE, Sundaramurthi JC, Salazar A, Gehrmann T, et al. Mycobacterium tuberculosis whole genome sequences from Southern India suggest novel resistance mechanisms and the need for region-specific diagnostics. Clin Infect Dis. 2017;64:1494–501.
Wollenberg KR, Desjardins CA, Zalutskaya A, Slodovnikova V, Oler AJ, Quiñones M, et al. Whole-genome sequencing of Mycobacterium tuberculosis provides insight into the evolution and genetic composition of drug-resistant tuberculosis in Belarus. J Clin Microbiol. 2017;55:457–69.
Koch A, Wilkinson RJ. The road to drug resistance in Mycobacterium tuberculosis . Genome Biol. 2014. https://doi.org/10.1186/s13059-014-0520-1 .
Walker TM, Lalor MK, Broda A, Saldana Ortega L, Morgan M, Parker L, et al. Assessment of Mycobacterium tuberculosis transmission in Oxfordshire, UK, 2007–12, with whole pathogen genome sequences: an observational study. Lancet Respir Med. 2014;2:285–92.
Gandhi NR, Moll A, Sturm AW, Pawinski R, Govender T, Lalloo U, et al. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–80.
Casali N, Nikolayevskyy V, Balabanova Y, Ignatyeva O, Kontsevaya I, Harris SR, et al. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 2012;22:735–45.
Guerra-Assunção JA, Houben RMGJ, Crampin AC, Mzembe T, Mallard K, Coll F, et al. Recurrence due to relapse or reinfection with Mycobacterium tuberculosis : a whole-genome sequencing approach in a large, population-based cohort with a high HIV infection prevalence and active follow-up. J Infect Dis. 2015;211:1154–63.
Perdigão J, Silva H, Machado D, Macedo R, Maltez F, Silva C, et al. Unraveling Mycobacterium tuberculosis genomic diversity and evolution in Lisbon, Portugal, a highly drug resistant setting. BMC Genomics. 2014. https://doi.org/10.1186/1471-2164-15-991 .
Lalor MK, Casali N, Walker TM, Anderson LF, Davidson JA, Ratna N, et al. The use of whole-genome sequencing in cluster investigation of a multidrug-resistant tuberculosis outbreak. Eur Respir J. 2018. https://doi.org/10.1183/13993003.02313-2017 .
Bjorn-Mortensen K, Soborg B, Koch A, Ladefoged K, Merker M, Lillebaek T, et al. Tracing Mycobacterium tuberculosis transmission by whole genome sequencing in a high incidence setting: a retrospective population-based study in East Greenland. Sci Rep. 2016. https://doi.org/10.1038/srep33180 .
Packer S, Green C, Brooks-Pollock E, Chaintarli K, Harrison S, Beck CR. Social network analysis and whole genome sequencing in a cohort study to investigate TB transmission in an educational setting. BMC Infect Dis. 2019. https://doi.org/10.1186/s12879-019-3734-8 .
Auld SC, Shah NS, Mathema B, Brown TS, Ismail N, Omar SV, et al. Extensively drug-resistant tuberculosis in South Africa: genomic evidence supporting transmission in communities. Eur Respir J. 2018. https://doi.org/10.1183/13993003.00246-2018 .
Ford C, Yusim K, Ioerger T, Feng S, Chase M, Greene M, et al. Mycobacterium tuberculosis —heterogeneity revealed through whole genome sequencing. Tuberculosis (Edinb). 2012;92:194–201.
Farmer P, Bayona J, Becerra M, Furin J, Henry C, Hiatt H, et al. The dilemma of MDR-TB in the global era. Int J Tuberc Lung Dis. 1998;2:869–76.
Xie YL, Chakravorty S, Armstrong DT, Hall SL, Via LE, Song T, et al. Evaluation of a rapid molecular drug-susceptibility test for tuberculosis. N Engl J Med. 2017;377:1043–54.
Lee RS, Behr MA. The implications of whole-genome sequencing in the control of tuberculosis. Ther Adv Infect Dis. 2016;3:47–62.
Jeanes C, O’Grady J. Diagnosing tuberculosis in the 21st century—Dawn of a genomics revolution? Int J Mycobacteriol. 2016;5:384–91.
Lee RS, Pai M. Real-time sequencing of Mycobacterium tuberculosis : are we there yet? J Clin Microbiol. 2017;55:1249–54.
Brown AC, Bryant JM, Einer-Jensen K, Holdstock J, Houniet DT, Chan JZM, et al. Rapid whole-genome sequencing of mycobacterium tuberculosis isolates directly from clinical samples. J Clin Microbiol. 2015;53:2230–7.
Doyle RM, Burgess C, Williams R, Gorton R, Booth H, Brown J, et al. Direct whole-genome sequencing of sputum accurately identifies drug-resistant Mycobacterium tuberculosis faster than MGIT culture sequencing. J Clin Microbiol. 2018. https://doi.org/10.1128/JCM.00666-18 .
Lowenthal P, Lin S-YG, Desmond E, Shah N, Flood J, Barry PM. Evaluation of the impact of a sequencing assay for detection of drug resistance on the clinical management of tuberculosis. Clin Infect Dis. 2018. https://doi.org/10.1093/cid/ciy937 .
Judge K, Harris SR, Reuter S, Parkhill J, Peacock SJ. Early insights into the potential of the Oxford Nanopore MinION for the detection of antimicrobial resistance genes. J Antimicrob Chemother. 2015;70:2775–8.
Schmidt K, Mwaigwisya S, Crossman LC, Doumith M, Munroe D, Pires C, et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother. 2017;72:104–14.
Cohen T, van Helden PD, Wilson D, Colijn C, Mclaughlin MM, Abubakar I, et al. Mixed-strain Mycobacterium tuberculosis infections and the implications for tuberculosis treatment and control. Clin Microbiol Rev. 2012;25:708–19.
World Health Organization. The use of next-generation sequencing technologies for the detection of mutations associated with drug resistance in Mycobacterium tuberculosis complex: technical guide. Geneva: WHO; 2018. https://apps.who.int/iris/bitstream/handle/10665/274443/WHO-CDS-TB-2018.19-eng.pdf?sequence=1&isAllowed=y . Accessed 9 July 2019
Walker TM, Cruz ALG, Peto TE, Smith EG, Esmail H, Crook DW. Tuberculosis is changing. Lancet Infect Dis. 2017;17:359–61.
Colman RE, Anderson J, Lemmer D, Lehmkuhl E, Georghiou SB, Heaton H, et al. Rapid drug susceptibility testing of drug-resistant Mycobacterium tuberculosis isolates directly from clinical samples by use of amplicon sequencing: a proof-of-concept study. J Clin Microbiol. 2016;54:2058–67.
Download references
Acknowledgements
The authors thank Arlin Keo for her assistance with the creation of Fig. 1 .
The authors’ work has been funded by the National Institute of Allergy and Infectious Diseases (grant number U19AI110818 to AME) and by The National Heart Lung and Blood Institute (K08 HL139994 to KAC) and the Burroughs Wellcome Fund CAMS (to KAC). The funders had no role in preparation of the manuscript.
Author information
Authors and affiliations.
Division of Pulmonary and Critical Care Medicine, Johns Hopkins University School of Medicine, Baltimore, MA, 21205, USA
Keira A. Cohen
Broad Institute of Harvard and Massachusetts Institute of Technology, 415 Main Street, Cambridge, MA, 02142, USA
Abigail L. Manson, Christopher A. Desjardins, Thomas Abeel & Ashlee M. Earl
Delft Bioinformatics Lab, Delft University of Technology, 2628, XE, Delft, The Netherlands
Thomas Abeel
You can also search for this author in PubMed Google Scholar
Contributions
KAC, ALM, CAD, TA, and AME wrote the manuscript. All authors read and approved the final manuscript.
Authors’ information
Keira A. Cohen, MD is a pulmonary and critical care medicine specialist at Johns Hopkins University School of Medicine. Abigail L. Manson, PhD is a senior computational biologist and computational group leader for Bacterial Genomics Group in the Infectious Disease and Microbiome Program at the Broad Institute of MIT and Harvard. Christopher Desjardins, PhD is a senior computational biologist working in the Infectious Disease and Microbiome Program at the Broad Institute of MIT and Harvard. Thomas Abeel, PhD is an assistant professor of bioinformatics at Delft University of Technology and a visiting scientist at the Broad Institute of MIT and Harvard. Ashlee M. Earl, PhD is a research scientist and senior group leader for the Bacterial Genomics Group in the Infectious Disease and Microbiome Program at the Broad Institute of MIT and Harvard.
Corresponding authors
Correspondence to Keira A. Cohen or Ashlee M. Earl .
Ethics declarations
Competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License ( http://creativecommons.org/licenses/by/4.0/ ), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated.
Reprints and permissions
About this article
Cite this article.
Cohen, K.A., Manson, A.L., Desjardins, C.A. et al. Deciphering drug resistance in Mycobacterium tuberculosis using whole-genome sequencing: progress, promise, and challenges. Genome Med 11 , 45 (2019). https://doi.org/10.1186/s13073-019-0660-8
Download citation
Published : 25 July 2019
DOI : https://doi.org/10.1186/s13073-019-0660-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Genome Medicine
ISSN: 1756-994X
- General enquiries: [email protected]

IMAGES
COMMENTS
We will be awarding a £100 first prize and two £50 runner-up prizes in both Medicine and Veterinary Medicine. There will also be an unlimited number of 'highly commended' prizes in both categories. Eligibility criteria This competition is open to Year 12 (S5 - Scotland, Y13 - N.I.) students attending a UK state
Newnham College of the University of Cambridge runs a medicine essay competition with a twist: Only female students are allowed to enter. Again, students have a choice of three differing questions. For example, the questions in the 2021-22 competition were:
Students in Year 12 (S5 - Scotland, Y13 - N.I.) attending a UK state school are invited to choose one question from either Medicine and Veterinary and submit an essay of between 1,000-1,500 words by the 6th June 2022. We will award one £100 first prize and two £50 second place prizes in both Medicine and Veterinary Medicine.
There are prizes available of £50 for first place, £30 for second place and £20 for third place. The deadline has varied from year-to-year: it was June in 2022 and April in 2023, so keep an eye on their website for more details. Learn about all of the competitions and prizes you can enter at school to boost your Medicine application.
Essay competitions for Year 12 students applying to medicine. Throughout the year, there are different essay competitions open for sixth form students to enter. We've compiled a list of some of the ones that are most relevant for your medicine application. Some of the deadlines are soon, whilst others are in a few months, so be selective and ...
The Woolf Essay Prize 2024 has now closed. Check back here in January 2025 for the 2025 competition! In 1928, Virginia Woolf addressed the Newnham Arts Society on the Subject of 'Women and Fiction', and from this talk emerged her seminal text, A Room of One's Own. Newnham is very proud of its place in the history of women's education ...
This essay competition is designed to give students the opportunity to develop and showcase their independent study and writing skills. Unfortunately, for external reasons, the essay won't be running in 2023, but may well be running in 2024 so do keep an eye out so you don't miss it! Sample Essay Questions from 2020.
Discourse, debate, and analysis Cambridge Re:think Essay Competition 2024 This year, CCIR saw over 4,200 submissions from more than 50 countries. Of these 4,200 essays, our jury panel, consists of scholars across the Atlantic, selected approximately 350 Honourable Mention students, and 33 award winners. The mission of the Re:think essay competition has always been to encourage critical […]
Destiny-Alliah Colombo. I am a Year 11 student who is interested in studying Maths, Chemistry and Biology at a higher level. I have been inspired to enter this essay competition due to my interest in reading medicine at University, and I have enjoyed developing my writing and researching skills through the process of formulating this essay.
John Fry prize. Deadline: Thursday 1 August 2024 Open to: Medical, nursing and allied healthcare students with an interest in general practice and primary care Apply here CAIPE John Horder Team Award and John Horder Student Award. Deadline: Wednesday 31 July 2024 Open to: Individuals or teams working within the community who can demonstrate outstanding principles of collaborative working and ...
The Future of Medicine. The Minds Underground™ Medicine Essay Competition is open to students in Year 12. The competition provides students with an opportunity to engage in university-level research, hone their writing & argumentative skills and prepare for university interviews. Entrants must choose 1 question to answer.
deferred 2023 entry. You do not have to be attending a school in the UK to enter this competition. Competition rules You may only submit one entry to the Cottrell Essay Prize for Medicine and Veterinary Medicine, and you must only answer one question from either Medicine or Veterinary Medicine. Your essay should be between 1,000-1,500 words.
Winner of the MedTech Foundation & Cambridge Medicine Journal Essay Competition 2021. 2021; Perspectives; Read more about What impact can simulations have in advancing medicine? Log in to post comments; More. Journal Issues. Show — Journal Issues Hide — Journal Issues. 2021; 2020; 2019; 2018; 2017; 2016; 2015; 2014; 2013; 2012;
Human papillomaviruses (HPV) are a large family of double-stranded DNA viruses that infect skin and mucosal cells [1]. Of the more than 100 recognised genotypes, at least 13 are considered to be oncogenic (or 'high-risk'). The two most common of these, i.e. type 16 and 18, are known to cause approximately 70 per cent of all cervical cancers.
All essay competitions and events at Cambridge (both online and in-person) can be found here 🔗 🌟. Magdalene College Arts and Humanities Essay Competition 2024 🔗 🌟 Any student in their penultimate year at a state school can enter this competition, which will open in early 2024. Last year, there were 12 questions covering a variety of ...
The following pages contain information about our Essay Prizes run for Lower and Upper 6th Students internationally, including how to apply. The Robson History Prize will not run in 2024 but we are expecting to run it again in 2025.
Medicine or law; medicine essay competitions; what does cambridge actually want from a medical applicant? stem extra curriculars/work experience (year 10) 2023 Immerse education essay competition; Will maths help with Cambridge Medicine; Great, I want to get into medicine. Now what? Immerse Education essay competition 2022; Advice on applying ...
Contents: Cities and Settlements The population of all cities and urban settlements in Omsk Oblast according to census results and latest official estimates. The icon links to further information about a selected place including its population structure (gender).
This year's essay prize questions get to the heart of some of the key issues affecting Medicine and Veterinary Medicine during this extraordinary time. Students in year 12 (or equivalent) attending schools/colleges in the UK and overseas are invited to choose one question from either Medicine or Veterinary Medicine and submit an essay of ...
Omsk State Medical University, located in Omsk, Siberia, is a top choice for studying MBBS in Russia. Founded in 1920 as the Medical Faculty of the Siberian Institute of Veterinary Medicine and Zoology, it offers a variety of undergraduate and postgraduate programs in different medical fields, including General Medicine, Pediatrics, Dentistry, Pharmacy, and Preventive Medicine.
2 Competition rules You may only submit one entry to the Cottrell Essay Prize for Medicine and Veterinary Medicine, and you must answer only one question from either a medical perspective, a veterinary perspective, or a perspective that considers both: Your essay should be between 1,000-1,500 words.This word limit includes footnotes and
This chapter presents history, economic statistics, and federal government directories of Omsk Oblast. Omsk Oblast is situated in the south of the Western Siberian Plain on the middle reaches of the Irtysh river.
Tuberculosis (TB) is a global infectious threat that is intensified by an increasing incidence of highly drug-resistant disease. Whole-genome sequencing (WGS) studies of Mycobacterium tuberculosis, the causative agent of TB, have greatly increased our understanding of this pathogen. Since the first M. tuberculosis genome was published in 1998, WGS has provided a more complete account of the ...