Breast Cancer Research


Breast Cancer Risk Factors
Breast Cancer Research is presenting our Retrospective Collection on "Breast Cancer Risk Factors." Celebrating 'Breast Cancer Awareness Month (1 October- 31 October)', with this Collection, we aim to gain valuable insights into the multifaceted aspects of breast cancer risk to promote awareness, prevention, and early detection.
NEW CROSS-JOURNAL COLLECTIONS Find out more by clicking the links below:
Artif icial Intelligence in Breast Imaging PDGFB in Br east Cancer Initiation,Progression, and Metastasis

Aims and scope
- Most accessed
Single-cell transcriptional atlas of tumor-associated macrophages in breast cancer
Authors: Yupeng Zhang, Fan Zhong and Lei Liu
Challenges and improvements in HER2 scoring and histologic evaluation: insights from a national proficiency testing scheme for breast cancer diagnosis in China
Authors: Xuemin Xue, Lei Guo, Changyuan Guo, Liwei Xu, Lin Li, Lin Yang, Xin Wang, Wei Rao, Pei Yuan, Jiali Mu, Jiangtao Li, Bingning Wang, Quan Zhou, Weicheng Xue, Fei Ma, Wenjing Yang…
Analysis of ductal carcinoma in situ by self-reported race reveals molecular differences related to outcome
Authors: Siri H. Strand, Kathleen E. Houlahan, Vernal Branch, Thomas Lynch, Belén Rivero-Guitiérrez, Bryan Harmon, Fergus Couch, Kristalyn Gallagher, Mark Kilgore, Shi Wei, Angela DeMichele, Tari King, Priscilla McAuliffe, Christina Curtis, Kouros Owzar, Jeffrey R. Marks…
Elevated expression of Aurora-A/ AURKA in breast cancer associates with younger age and aggressive features
Authors: L. M. Ingebriktsen, R. O. C. Humlevik, A. A. Svanøe, A. K. M. Sæle, I. Winge, K. Toska, M. B. Kalvenes, B. Davidsen, A. Heie, G. Knutsvik, C. Askeland, I. M. Stefansson, E. A. Hoivik, L. A. Akslen and E. Wik
Systematic assessment of HER2 status in ductal carcinoma in situ of the breast: a perspective on the potential clinical relevance
Authors: Mieke R. Van Bockstal, Jelle Wesseling, Ester H. Lips, Marjolein Smidt, Christine Galant and Carolien H. M. van Deurzen
Most recent articles RSS
View all articles
Serum thymidine kinase 1 activity as a pharmacodynamic marker of cyclin-dependent kinase 4/6 inhibition in patients with early-stage breast cancer receiving neoadjuvant palbociclib
Authors: Nusayba Bagegni, Shana Thomas, Ning Liu, Jingqin Luo, Jeremy Hoog, Donald W. Northfelt, Matthew P. Goetz, Andres Forero, Mattias Bergqvist, Jakob Karen, Magnus Neumüller, Edward M. Suh, Zhanfang Guo, Kiran Vij, Souzan Sanati, Matthew Ellis…
Choosing the right cell line for breast cancer research
Authors: Deborah L Holliday and Valerie Speirs
Triple-negative breast cancer molecular subtyping and treatment progress
Authors: Li Yin, Jiang-Jie Duan, Xiu-Wu Bian and Shi-cang Yu
Breast asymmetry and predisposition to breast cancer
Authors: Diane Scutt, Gillian A Lancaster and John T Manning
Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer
Authors: Suzanne A Eccles, Eric O Aboagye, Simak Ali, Annie S Anderson, Jo Armes, Fedor Berditchevski, Jeremy P Blaydes, Keith Brennan, Nicola J Brown, Helen E Bryant, Nigel J Bundred, Joy M Burchell, Anna M Campbell, Jason S Carroll, Robert B Clarke, Charlotte E Coles…
Most accessed articles RSS

Editor-in-Chief
Lewis Chodosh , University of Pennsylvania, USA

Trending in the Media
Click here to see the most popular articles published in Breast Cancer Research in the past three months.

BCR's 20th Anniversary
20 years ago Breast Cancer Research published its first articles with BMC. Well-respected in the field, the journal has continually placed in the first quartile of the ‘Oncology’ category of Journal Citation Reports. Over the past decade, Breast Cancer Research (BCR) has also become the highest ranked breast cancer focused title in the field.
Look back at the journal’s milestone achievements and article highlights .

Featured Review - Artificial intelligence in mammographic phenotyping of breast cancer risk: a narrative review
In this review, we provide a useful reference for AI researchers investigating image-based breast cancer risk assessment while indicating key priorities and challenges that, if properly addressed, could accelerate the implementation of AI-assisted risk stratification to future refine and individualize breast cancer screening strategies.
Springer Nature Oncology Portfolio
Discover the range of academic oncology titles at Springer Nature here .

Article Menu

- Subscribe SciFeed
- Recommended Articles
- Google Scholar
- on Google Scholar
- Table of Contents
Find support for a specific problem in the support section of our website.
Please let us know what you think of our products and services.
Visit our dedicated information section to learn more about MDPI.
JSmol Viewer
Single-cell rna-sequencing: opening new horizons for breast cancer research.

1. Introduction
2. the functional principle and workflow of scrna-seq, 2.1. single-cell isolation, 2.2. reverse transcription, amplification, and sequencing, 2.2.1. pcr after polya tailing, 2.2.2. template-switching-based pcr, 2.2.3. in vitro transcription (ivt), 2.3. data analysis, 3. application of scrna-seq in breast cancer, 3.1. application of scrna-seq in exploring the heterogeneity of breast cancer, 3.2. application of scrna-seq in tme of breast cancer, 3.3. application of scrna-seq in therapy of breast cancer, 3.4. application of scrna-seq in drug resistance of breast cancer, 3.5. application of scrna-seq in metastasis of breast cancer, 4. potential future directions of scrna-seq in breast cancer research, 5. conclusions, author contributions, conflicts of interest.
- Jiang, G.; Tu, J.; Zhou, L.; Dong, M.; Fan, J.; Chang, Z.; Zhang, L.; Bian, X.; Liu, S. Single-cell transcriptomics reveal the heterogeneity and dynamic of cancer stem-like cells during breast tumor progression. Cell Death Dis. 2021 , 12 , 979. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Wilkinson, L.; Gathani, T. Understanding breast cancer as a global health concern. Br. J. Radiol. 2022 , 95 , 20211033. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017 , 4 , 227. [ Google Scholar ] [ CrossRef ]
- Tuasha, N.; Petros, B. Heterogeneity of Tumors in Breast Cancer: Implications and Prospects for Prognosis and Therapeutics. Scientifica 2020 , 2020 , 4736091. [ Google Scholar ] [ CrossRef ]
- Luond, F.; Tiede, S.; Christofori, G. Breast cancer as an example of tumour heterogeneity and tumour cell plasticity during malignant progression. Br. J. Cancer 2021 , 125 , 164–175. [ Google Scholar ] [ CrossRef ]
- Thakur, S.; Haider, S.; Natrajan, R. Implications of tumour heterogeneity on cancer evolution and therapy resistance: Lessons from breast cancer. J. Pathol. 2023 , 260 , 621–636. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Qian, J.; Olbrecht, S.; Boeckx, B.; Vos, H.; Laoui, D.; Etlioglu, E.; Wauters, E.; Pomella, V.; Verbandt, S.; Busschaert, P.; et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020 , 30 , 745–762. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Xiao, Y.; Yu, D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol. Ther. 2021 , 221 , 107753. [ Google Scholar ] [ CrossRef ]
- Deepak, K.G.K.; Vempati, R.; Nagaraju, G.P.; Dasari, V.R.; Nagini, S.; Rao, D.N.; Malla, R.R. Tumor microenvironment: Challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol. Res. 2020 , 153 , 104683. [ Google Scholar ] [ CrossRef ]
- Manohar, S.M.; Shah, P.; Nair, A. Flow cytometry: Principles, applications and recent advances. Bioanalysis 2021 , 13 , 181–198. [ Google Scholar ] [ CrossRef ]
- Liu, C.C.; Steen, C.B.; Newman, A.M. Computational approaches for characterizing the tumor immune microenvironment. Immunology 2019 , 158 , 70–84. [ Google Scholar ] [ CrossRef ]
- Fincham, R.E.A.; Bashiri, H.; Lau, M.C.; Yeong, J. Editorial: Multiplex Immunohistochemistry/Immunofluorescence Technique: The Potential and Promise for Clinical Application. Front. Mol. Biosci. 2022 , 9 , 831383. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Yamada, S.; Nomura, S. Review of Single-Cell RNA Sequencing in the Heart. Int. J. Mol. Sci. 2020 , 21 , 8345. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Wang, Y.; Mashock, M.; Tong, Z.; Mu, X.; Chen, H.; Zhou, X.; Zhang, H.; Zhao, G.; Liu, B.; Li, X. Changing Technologies of RNA Sequencing and Their Applications in Clinical Oncology. Front. Oncol. 2020 , 10 , 447. [ Google Scholar ] [ CrossRef ]
- Kulkarni, A.; Anderson, A.G.; Merullo, D.P.; Konopka, G. Beyond bulk: A review of single cell transcriptomics methodologies and applications. Curr. Opin. Biotechnol. 2019 , 58 , 129–136. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Eberwine, J.; Yeh, H.; Miyashiro, K.; Cao, Y.; Nair, S.; Finnell, R.; Zettel, M.; Coleman, P. Analysis of gene expression in single live neurons. Proc. Natl. Acad. Sci. USA 1992 , 89 , 3010–3014. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Zhang, J.; Song, C.; Tian, Y.; Yang, X. Single-Cell RNA Sequencing in Lung Cancer: Revealing Phenotype Shaping of Stromal Cells in the Microenvironment. Front. Immunol. 2021 , 12 , 802080. [ Google Scholar ] [ CrossRef ]
- Ding, S.; Chen, X.; Shen, K. Single-cell RNA sequencing in breast cancer: Understanding tumor heterogeneity and paving roads to individualized therapy. Cancer Commun. 2020 , 40 , 329–344. [ Google Scholar ] [ CrossRef ]
- Lei, Y.; Tang, R.; Xu, J.; Wang, W.; Zhang, B.; Liu, J.; Yu, X.; Shi, S. Applications of single-cell sequencing in cancer research: Progress and perspectives. J. Hematol. Oncol. 2021 , 14 , 91. [ Google Scholar ] [ CrossRef ]
- Luecken, M.D.; Theis, F.J. Current best practices in single-cell RNA-seq analysis: A tutorial. Mol. Syst. Biol. 2019 , 15 , e8746. [ Google Scholar ] [ CrossRef ]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009 , 6 , 377–382. [ Google Scholar ] [ CrossRef ]
- Pensold, D.; Zimmer-Bensch, G. Methods for Single-Cell Isolation and Preparation. In Single-Cell Sequencing and Methylation: Methods and Clinical Applications ; Yu, B., Zhang, J., Zeng, Y., Li, L., Wang, X., Eds.; Springer: Singapore, 2020; pp. 7–27. [ Google Scholar ]
- Foley, J.W.; Zhu, C.; Jolivet, P.; Zhu, S.X.; Lu, P.; Meaney, M.J.; West, R.B. Gene expression profiling of single cells from archival tissue with laser-capture microdissection and Smart-3SEQ. Genome Res. 2019 , 29 , 1816–1825. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Aguilar-Bravo, B.; Sancho-Bru, P. Laser capture microdissection: Techniques and applications in liver diseases. Hepatol. Int. 2019 , 13 , 138–147. [ Google Scholar ] [ CrossRef ]
- Rao, B.H.; Soucek, P.; Hlavac, V. Laser Capture Microdissection: A Gear for Pancreatic Cancer Research. Int. J. Mol. Sci. 2022 , 23 , 14566. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Kolodziejczyk, A.A.; Kim, J.K.; Svensson, V.; Marioni, J.C.; Teichmann, S.A. The technology and biology of single-cell RNA sequencing. Mol. Cell 2015 , 58 , 610–620. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Xu, X.; Wang, J.; Wu, L.; Guo, J.; Song, Y.; Tian, T.; Wang, W.; Zhu, Z.; Yang, C. Microfluidic Single-Cell Omics Analysis. Small 2020 , 16 , e1903905. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Zhang, Q.; Gu, M.L. Single-cell sequencing and its application in breast cancer. Yi Chuan 2020 , 42 , 250–268. [ Google Scholar ]
- Tung, P.Y.; Blischak, J.D.; Hsiao, C.J.; Knowles, D.A.; Burnett, J.E.; Pritchard, J.K.; Gilad, Y. Batch effects and the effective design of single-cell gene expression studies. Sci. Rep. 2017 , 7 , 39921. [ Google Scholar ] [ CrossRef ]
- El-Hajjar, L.; Ali, A.F.; Nasr, R.A. Guide to Flow Cytometry: Components, Basic Principles, Experimental Design, and Cancer Research Applications. Curr. Protoc. 2023 , 3 , e721. [ Google Scholar ] [ CrossRef ]
- Ellsworth, D.L.; Blackburn, H.L.; Shriver, C.D.; Rabizadeh, S.; Soon-Shiong, P.; Ellsworth, R.E. Single-cell sequencing and tumorigenesis: Improved understanding of tumor evolution and metastasis. Clin. Transl. Med. 2017 , 6 , 15. [ Google Scholar ] [ CrossRef ]
- Sasagawa, Y.; Nikaido, I.; Hayashi, T.; Danno, H.; Uno, K.D.; Imai, T.; Ueda, H.R. Quartz-Seq: A highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol. 2013 , 14 , R31. [ Google Scholar ] [ CrossRef ]
- Sasagawa, Y.; Danno, H.; Takada, H.; Ebisawa, M.; Tanaka, K.; Hayashi, T.; Kurisaki, A.; Nikaido, I. Quartz-Seq2: A high-throughput single-cell RNA-sequencing method that effectively uses limited sequence reads. Genome Biol. 2018 , 19 , 29. [ Google Scholar ] [ CrossRef ]
- Fan, X.; Zhang, X.; Wu, X.; Guo, H.; Hu, Y.; Tang, F.; Huang, Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015 , 16 , 148. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Sheng, K.; Zong, C. Single-Cell RNA-Seq by Multiple Annealing and Tailing-Based Quantitative Single-Cell RNA-Seq (MATQ-Seq). Methods Mol. Biol. 2019 , 1979 , 57–71. [ Google Scholar ] [ PubMed ]
- Sheng, K.; Cao, W.; Niu, Y.; Deng, Q.; Zong, C. Effective detection of variation in single-cell transcriptomes using MATQ-seq. Nat. Methods 2017 , 14 , 267–270. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Picelli, S.; Björklund, Å.K.; Faridani, O.R.; Sagasser, S.; Winberg, G.; Sandberg, R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 2013 , 10 , 1096–1098. [ Google Scholar ] [ CrossRef ]
- Picelli, S.; Faridani, O.R.; Björklund, Å.K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014 , 9 , 171–181. [ Google Scholar ] [ CrossRef ]
- Ramsköld, D.; Luo, S.; Wang, Y.C.; Li, R.; Deng, Q.; Faridani, O.R.; Daniels, G.A.; Khrebtukova, I.; Loring, J.F.; Laurent, L.C.; et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012 , 30 , 777–782. [ Google Scholar ] [ CrossRef ]
- Hagemann-Jensen, M.; Ziegenhain, C.; Chen, P.; Ramsköld, D.; Hendriks, G.J.; Larsson, A.J.; Faridani, O.R.; Sandberg, R. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 2020 , 38 , 708–714. [ Google Scholar ] [ CrossRef ]
- Hahaut, V.; Pavlinic, D.; Carbone, W.; Schuierer, S.; Balmer, P.; Quinodoz, M.; Renner, M.; Roma, G.; Cowan, C.S.; Picelli, S. Fast and highly sensitive full-length single-cell RNA sequencing using FLASH-seq. Nat. Biotechnol. 2023 , 40 , 1447–1451. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Natarajan, K.N. Single-Cell Tagged Reverse Transcription (STRT-Seq). Methods Mol. Biol. 2019 , 1979 , 133–153. [ Google Scholar ] [ PubMed ]
- Hochgerner, H.; Lönnerberg, P.; Hodge, R.; Mikes, J.; Heskol, A.; Hubschle, H.; Lin, P.; Picelli, S.; La Manno, G.; Ratz, M.; et al. STRT-seq-2i: Dual-index 5′ single cell and nucleus RNA-seq on an addressable microwell array. Sci. Rep. 2017 , 7 , 16327. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Ni, J.; Hu, C.; Li, H.; Li, X.; Fu, Q.; Czajkowsky, D.M.; Guo, Y.; Shao, Z. Significant improvement in data quality with simplified SCRB-seq. Acta Biochim. Biophys. Sin. 2020 , 52 , 457–459. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015 , 161 , 1202–1214. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Bageritz, J.; Raddi, G. Single-Cell RNA Sequencing with Drop-Seq. Methods Mol. Biol. 2019 , 1979 , 73–85. [ Google Scholar ] [ PubMed ]
- Shapiro, E.; Biezuner, T.; Linnarsson, S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 2013 , 14 , 618–630. [ Google Scholar ] [ CrossRef ]
- Picelli, S. Single-cell RNA-sequencing: The future of genome biology is now. RNA Biol. 2017 , 14 , 637–650. [ Google Scholar ] [ CrossRef ]
- Dal Molin, A.; Di Camillo, B. How to design a single-cell RNA-sequencing experiment: Pitfalls, challenges and perspectives. Brief. Bioinform. 2019 , 20 , 1384–1394. [ Google Scholar ] [ CrossRef ]
- Zong, C.; Lu, S.; Chapman, A.R.; Xie, X.S. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 2012 , 338 , 1622–1626. [ Google Scholar ] [ CrossRef ]
- Grun, D.; van Oudenaarden, A. Design and Analysis of Single-Cell Sequencing Experiments. Cell 2015 , 163 , 799–810. [ Google Scholar ] [ CrossRef ]
- Hashimshony, T.; Wagner, F.; Sher, N.; Yanai, I. CEL-Seq: Single-cell RNA-Seq by multiplexed linear amplification. Cell. Rep. 2012 , 2 , 666–673. [ Google Scholar ] [ CrossRef ]
- Hashimshony, T.; Senderovich, N.; Avital, G.; Klochendler, A.; De Leeuw, Y.; Anavy, L.; Gennert, D.; Li, S.; Livak, K.J.; Rozenblatt-Rosen, O.; et al. CEL-Seq2: Sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016 , 17 , 77. [ Google Scholar ] [ CrossRef ]
- Yanai, I.; Hashimshony, T. CEL-Seq2-Single-Cell RNA Sequencing by Multiplexed Linear Amplification. Methods Mol. Biol. 2019 , 1979 , 45–56. [ Google Scholar ]
- Jaitin, D.A.; Kenigsberg, E.; Keren-Shaul, H.; Elefant, N.; Paul, F.; Zaretsky, I.; Mildner, A.; Cohen, N.; Jung, S.; Tanay, A.; et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 2014 , 343 , 776–779. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Keren-Shaul, H.; Kenigsberg, E.; Jaitin, D.A.; David, E.; Paul, F.; Tanay, A.; Amit, I. MARS-seq2.0: An experimental and analytical pipeline for indexed sorting combined with single-cell RNA sequencing. Nat. Protoc. 2019 , 14 , 1841–1862. [ Google Scholar ] [ CrossRef ]
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015 , 161 , 1187–1201. [ Google Scholar ] [ CrossRef ]
- Zappia, L.; Phipson, B.; Oshlack, A. Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database. PLoS Comput. Biol. 2018 , 14 , e1006245. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018 , 36 , 411–420. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- McCarthy, D.J.; Campbell, K.R.; Lun, A.T.; Wills, Q.F. Scater: Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 2017 , 33 , 1179–1186. [ Google Scholar ] [ CrossRef ]
- Wolf, F.A.; Angerer, P.; Theis, F.J. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 2018 , 19 , 15. [ Google Scholar ] [ CrossRef ]
- Slovin, S.; Carissimo, A.; Panariello, F.; Grimaldi, A.; Bouché, V.; Gambardella, G.; Cacchiarelli, D. Single-Cell RNA Sequencing Analysis: A Step-by-Step Overview. Methods Mol. Biol. 2021 , 2284 , 343–365. [ Google Scholar ] [ PubMed ]
- Su, M.; Pan, T.; Chen, Q.Z.; Zhou, W.W.; Gong, Y.; Xu, G.; Yan, H.Y.; Li, S.; Shi, Q.Z.; Zhang, Y.; et al. Data analysis guidelines for single-cell RNA-seq in biomedical studies and clinical applications. Mil. Med. Res. 2022 , 9 , 68. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Ding, J.; Sharon, N.; Bar-Joseph, Z. Temporal modelling using single-cell transcriptomics. Nat. Rev. Genet. 2022 , 23 , 355–368. [ Google Scholar ] [ CrossRef ]
- Cheng, C.; Chen, W.; Jin, H.; Chen, X. A Review of Single-Cell RNA-Seq Annotation, Integration, and Cell-Cell Communication. Cells 2023 , 12 , 1970. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Muciño-Olmos, E.A.; Vázquez-Jiménez, A.; Avila-Ponce de León, U.; Matadamas-Guzman, M.; Maldonado, V.; López-Santaella, T.; Hernández-Hernández, A.; Resendis-Antonio, O. Unveiling functional heterogeneity in breast cancer multicellular tumor spheroids through single-cell RNA-seq. Sci. Rep. 2020 , 10 , 12728. [ Google Scholar ] [ CrossRef ]
- Gray, G.K.; Li, C.M.C.; Rosenbluth, J.M.; Selfors, L.M.; Girnius, N.; Lin, J.R.; Schackmann, R.C.; Goh, W.L.; Moore, K.; Shapiro, H.K.; et al. A human breast atlas integrating single-cell proteomics and transcriptomics. Dev. Cell 2022 , 57 , 1400–1420. [ Google Scholar ] [ CrossRef ]
- Kumar, T.; Nee, K.; Wei, R.; He, S.; Nguyen, Q.H.; Bai, S.; Blake, K.; Pein, M.; Gong, Y.; Sei, E. A spatially resolved single-cell genomic atlas of the adult human breast. Nature 2023 , 620 , 181–191. [ Google Scholar ] [ CrossRef ]
- Liu, S.Q.; Gao, Z.J.; Wu, J.; Zheng, H.M.; Li, B.; Sun, S.; Meng, X.Y.; Wu, Q. Single-cell and spatially resolved analysis uncovers cell heterogeneity of breast cancer. J. Hematol. Oncol. 2022 , 15 , 19. [ Google Scholar ] [ CrossRef ]
- Wu, S.Z.; Al-Eryani, G.; Roden, D.L.; Junankar, S.; Harvey, K.; Andersson, A.; Thennavan, A.; Wang, C.; Torpy, J.R.; Bartonicek, N.; et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 2021 , 53 , 1334–1347. [ Google Scholar ] [ CrossRef ]
- Liu, Q.; Wang, X. Classification of triple-negative breast cancer based on pathway enrichment levels. Med. Oncol. 2023 , 40 , 157. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Hu, X.E.; Yang, P.; Chen, S.; Wei, G.; Yuan, L.; Yang, Z.; Gong, L.; He, L.; Yang, L.; Peng, S.; et al. Clinical and biological heterogeneities in triple-negative breast cancer reveals a non-negligible role of HER2-low. Breast Cancer Res. 2023 , 25 , 34. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Hou, J.; Liu, W.; Yan, M.; Ren, Y.; Qian, C.; Fu, Y.; Wang, H.; Li, Z. Unveiling heterogeneity and prognostic markers in ductal breast cancer through single-cell RNA-seq. Cancer Cell Int. 2024 , 24 , 266. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Karaayvaz, M.; Cristea, S.; Gillespie, S.M.; Patel, A.P.; Mylvaganam, R.; Luo, C.C.; Specht, M.C.; Bernstein, B.E.; Michor, F.; Ellisen, L.W. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat. Commun. 2018 , 9 , 3588. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Chaffer, C.L.; Weinberg, R.A. Cancer cell of origin: Spotlight on luminal progenitors. Cell Stem Cell 2010 , 7 , 271–272. [ Google Scholar ] [ CrossRef ]
- Chung, W.; Eum, H.H.; Lee, H.O.; Lee, K.M.; Lee, H.B.; Kim, K.T.; Ryu, H.S.; Kim, S.; Lee, J.E.; Park, Y.H.; et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 2017 , 8 , 15081. [ Google Scholar ] [ CrossRef ]
- Azizi, E.; Carr, A.J.; Plitas, G.; Cornish, A.E.; Konopacki, C.; Prabhakaran, S.; Nainys, J.; Wu, K.; Kiseliovas, V.; Setty, M.; et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 2018 , 174 , 1293–1308. [ Google Scholar ] [ CrossRef ]
- Savas, P.; Virassamy, B.; Ye, C.; Salim, A.; Mintoff, C.P.; Caramia, F.; Salgado, R.; Byrne, D.J.; Teo, Z.L.; Dushyanthen, S.; et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 2018 , 24 , 986–993. [ Google Scholar ] [ CrossRef ]
- Ding, S.; Qiao, N.; Zhu, Q.; Tong, Y.; Wang, S.; Chen, X.; Tian, Q.; Xiao, Y.; Shen, K. Single-cell atlas reveals a distinct immune profile fostered by T cell-B cell crosstalk in triple negative breast cancer. Cancer Commun. 2023 , 43 , 661–684. [ Google Scholar ] [ CrossRef ]
- Rebuffet, L.; Melsen, J.E.; Escalière, B.; Basurto-Lozada, D.; Bhandoola, A.; Björkström, N.K.; Bryceson, Y.T.; Castriconi, R.; Cichocki, F.; Colonna, M.; et al. High-dimensional single-cell analysis of human natural killer cell heterogeneity. Nat. Immunol. 2024 , 25 , 1474–1488. [ Google Scholar ] [ CrossRef ]
- Mao, J.; Liu, L.L.; Shen, Q.; Cen, M. Integrating single-cell transcriptomics and machine learning to predict breast cancer prognosis: A study based on natural killer cell-related genes. J. Cell. Mol. Med. 2024 , 28 , e18549. [ Google Scholar ] [ CrossRef ]
- Xu, M.; Zhang, T.; Xia, R.; Wei, Y.; Wei, X. Targeting the tumor stroma for cancer therapy. Mol. Cancer 2022 , 21 , 208. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Ma, C.; Yang, C.; Peng, A.; Sun, T.; Ji, X.; Mi, J.; Wei, L.; Shen, S.; Feng, Q. Pan-cancer spatially resolved single-cell analysis reveals the crosstalk between cancer-associated fibroblasts and tumor microenvironment. Mol. Cancer 2023 , 22 , 170. [ Google Scholar ] [ CrossRef ]
- Cords, L.; Tietscher, S.; Anzeneder, T.; Langwieder, C.; Rees, M.; de Souza, N.; Bodenmiller, B. Cancer-associated fibroblast classification in single-cell and spatial proteomics data. Nat. Commun. 2023 , 14 , 4294. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Ning, L.; Quan, C.; Wang, Y.; Wu, Z.; Yuan, P.; Xie, N. scRNA-seq characterizing the heterogeneity of fibroblasts in breast cancer reveals a novel subtype SFRP4(+) CAF that inhibits migration and predicts prognosis. Front. Oncol. 2024 , 14 , 1348299. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Croizer, H.; Mhaidly, R.; Kieffer, Y.; Gentric, G.; Djerroudi, L.; Leclere, R.; Pelon, F.; Robley, C.; Bohec, M.; Meng, A.; et al. Deciphering the spatial landscape and plasticity of immunosuppressive fibroblasts in breast cancer. Nat. Commun. 2024 , 15 , 2806. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Houthuijzen, J.M.; De Bruijn, R.; Van Der Burg, E.; Drenth, A.P.; Wientjens, E.; Filipovic, T.; Bullock, E.; Brambillasca, C.S.; Pulver, E.M.; Nieuwland, M.; et al. CD26-negative and CD26-positive tissue-resident fibroblasts contribute to functionally distinct CAF subpopulations in breast cancer. Nat. Commun. 2023 , 14 , 183. [ Google Scholar ] [ CrossRef ]
- Gambardella, G.; Viscido, G.; Tumaini, B.; Isacchi, A.; Bosotti, R.; Di Bernardo, D. A single-cell analysis of breast cancer cell lines to study tumour heterogeneity and drug response. Nat. Commun. 2022 , 13 , 1714. [ Google Scholar ] [ CrossRef ]
- Huang, J.; Zhang, J.L.; Ang, L.; Li, M.C.; Zhao, M.; Wang, Y.; Wu, Q. Proposing a novel molecular subtyping scheme for predicting distant recurrence-free survival in breast cancer post-neoadjuvant chemotherapy with close correlation to metabolism and senescence. Front. Endocrinol. 2023 , 14 , 1265520. [ Google Scholar ] [ CrossRef ]
- Zhang, X.; Feng, R.; Guo, J.; Pan, L.; Yao, Y.; Gao, J. Integrated single-cell and bulk RNA sequencing analysis identifies a neoadjuvant chemotherapy-related gene signature for predicting survival and therapy in breast cancer. BMC Med. Genom. 2023 , 16 , 300. [ Google Scholar ] [ CrossRef ]
- Mei, J.; Cai, Y.; Chen, L.; Wu, Y.; Liu, J.; Qian, Z.; Jiang, Y.; Zhang, P.; Xia, T.; Pan, X.; et al. The heterogeneity of tumour immune microenvironment revealing the CRABP2/CD69 signature discriminates distinct clinical outcomes in breast cancer. Br. J. Cancer 2023 , 129 , 1645–1657. [ Google Scholar ] [ CrossRef ]
- Deng, J.; Thennavan, A.; Shah, S.; Bagdatlioglu, E.; Klar, N.; Heguy, A.; Marier, C.; Meyn, P.; Zhang, Y.; Labbe, K.; et al. Serial single-cell profiling analysis of metastatic TNBC during Nab-paclitaxel and pembrolizumab treatment. Breast Cancer Res. Treat. 2021 , 185 , 85–94. [ Google Scholar ] [ CrossRef ]
- Zhang, Y.; Chen, H.; Mo, H.; Hu, X.; Gao, R.; Zhao, Y.; Liu, B.; Niu, L.; Sun, X.; Yu, X.; et al. Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell 2021 , 39 , 1578–1593. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Brady, S.W.; McQuerry, J.A.; Qiao, Y.; Piccolo, S.R.; Shrestha, G.; Jenkins, D.F.; Layer, R.M.; Pedersen, B.S.; Miller, R.H.; Esch, A.; et al. Combating subclonal evolution of resistant cancer phenotypes. Nat. Commun. 2017 , 8 , 1231. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Ren, L.; Li, J.; Wang, C.; Lou, Z.; Gao, S.; Zhao, L.; Wang, S.; Chaulagain, A.; Zhang, M.; Li, X.; et al. Single cell RNA sequencing for breast cancer: Present and future. Cell Death Discov. 2021 , 7 , 104. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Kim, C.; Gao, R.; Sei, E.; Brandt, R.; Hartman, J.; Hatschek, T.; Crosetto, N.; Foukakis, T.; Navin, N.E. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell 2018 , 173 , 879–893. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Shaath, H.; Vishnubalaji, R.; Elango, R.; Khattak, S.; Alajez, N.M. Single-cell long noncoding RNA (lncRNA) transcriptome implicates MALAT1 in triple-negative breast cancer (TNBC) resistance to neoadjuvant chemotherapy. Cell Death Discov. 2021 , 7 , 23. [ Google Scholar ] [ CrossRef ]
- Prieto-Vila, M.; Usuba, W.; Takahashi, R.U.; Shimomura, I.; Sasaki, H.; Ochiya, T.; Yamamoto, Y. Single-Cell Analysis Reveals a Preexisting Drug-Resistant Subpopulation in the Luminal Breast Cancer Subtype. Cancer Res. 2019 , 79 , 4412–4425. [ Google Scholar ] [ CrossRef ]
- Gao, Y.; Li, X.; Zeng, C.; Liu, C.; Hao, Q.; Li, W.; Zhang, K.; Zhang, W.; Wang, S.; Zhao, H.; et al. CD63(+) Cancer-Associated Fibroblasts Confer Tamoxifen Resistance to Breast Cancer Cells through Exosomal miR-22. Adv. Sci. 2020 , 7 , 2002518. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Du, R.; Zhang, X.; Lu, X.; Ma, X.; Guo, X.; Shi, C.; Ren, X.; Ma, X.; He, Y.; Gao, Y.; et al. PDPN positive CAFs contribute to HER2 positive breast cancer resistance to trastuzumab by inhibiting antibody-dependent NK cell-mediated cytotoxicity. Drug Resist. Updates 2023 , 68 , 100947. [ Google Scholar ] [ CrossRef ]
- Gao, Y.; Wu, N.; Wang, S.; Yang, X.; Wang, X.; Xu, B. Concurrent mutations associated with trastuzumab-resistance revealed by single cell sequencing. Breast Cancer Res. Treat. 2021 , 187 , 613–624. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Yi, M.; Niu, M.; Wu, Y.; Ge, H.; Jiao, D.; Zhu, S.; Zhang, J.; Yan, Y.; Zhou, P.; Chu, Q.; et al. Combination of oral STING agonist MSA-2 and anti-TGF-beta/PD-L1 bispecific antibody YM101: A novel immune cocktail therapy for non-inflamed tumors. J. Hematol. Oncol. 2022 , 15 , 142. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Xu, K.; Wang, R.; Xie, H.; Hu, L.; Wang, C.; Xu, J.; Zhu, C.; Liu, Y.; Gao, F.; Li, X.; et al. Single-cell RNA sequencing reveals cell heterogeneity and transcriptome profile of breast cancer lymph node metastasis. Oncogenesis 2021 , 10 , 66. [ Google Scholar ] [ CrossRef ]
- Xu, K.; Zhang, W.; Wang, C.; Hu, L.; Wang, R.; Wang, C.; Tang, L.; Zhou, G.; Zou, B.; Xie, H.; et al. Integrative analyses of scRNA-seq and scATAC-seq reveal CXCL14 as a key regulator of lymph node metastasis in breast cancer. Hum. Mol. Genet. 2021 , 30 , 370–380. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Liu, Y.M.; Ge, J.Y.; Chen, Y.F.; Liu, T.; Chen, L.; Liu, C.C.; Ma, D.; Chen, Y.Y.; Cai, Y.W.; Xu, Y.Y.; et al. Combined Single-Cell and Spatial Transcriptomics Reveal the Metabolic Evolvement of Breast Cancer during Early Dissemination. Adv. Sci. 2023 , 10 , e2205395. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Bieniasz-Krzywiec, P.; Martín-Pérez, R.; Ehling, M.; García-Caballero, M.; Pinioti, S.; Pretto, S.; Kroes, R.; Aldeni, C.; Di Matteo, M.; Prenen, H.; et al. Podoplanin-Expressing Macrophages Promote Lymphangiogenesis and Lymphoinvasion in Breast Cancer. Cell Metab. 2019 , 30 , 917–936. [ Google Scholar ] [ CrossRef ]
- Shen, J.; Ma, H.; Chen, Y.; Shen, J. ScRNA-seq reveals the correlation between M2 phenotype of tumor-associated macrophages and lymph node metastasis of breast cancer. Oncol. Res. 2023 , 31 , 955–966. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Sanjaya, A.; Ratnawati, H.; Adhika, O.A.; Rahmatilah, F.R. The heterogeneity of breast cancer metastasis: A bioinformatics analysis utilizing single-cell RNA sequencing data. Breast Cancer Res. Treat. 2024 . [ Google Scholar ] [ CrossRef ]
- Sun, H.; Zhang, L.; Wang, Z.; Gu, D.; Zhu, M.; Cai, Y.; Li, L.; Tang, J.; Huang, B.; Bosco, B.; et al. Single-cell transcriptome analysis indicates fatty acid metabolism-mediated metastasis and immunosuppression in male breast cancer. Nat. Commun. 2023 , 14 , 5590. [ Google Scholar ] [ CrossRef ]
- Gulati, G.S.; D’Silva, J.P.; Liu, Y.; Wang, L.; Newman, A.M. Profiling cell identity and tissue architecture with single-cell and spatial transcriptomics. Nat. Rev. Mol. Cell Biol. 2024 . [ Google Scholar ] [ CrossRef ]
- Stein, C.M.; Weiskirchen, R.; Damm, F.; Strzelecka, P.M. Single-cell omics: Overview, analysis, and application in biomedical science. J. Cell. Biochem. 2021 , 122 , 1571–1578. [ Google Scholar ] [ CrossRef ]
- Srinivasan, S.; Leshchyk, A.; Johnson, N.T.; Korkin, D. A hybrid deep clustering approach for robust cell type profiling using single-cell RNA-seq data. RNA 2020 , 26 , 1303–1319. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Wang, J.; Chen, Y.; Zou, Q. Inferring gene regulatory network from single-cell transcriptomes with graph autoencoder model. PLoS Genet. 2023 , 19 , e1010942. [ Google Scholar ] [ CrossRef ] [ PubMed ]
Click here to enlarge figure
| Method | RNA-Capture | Transcript Coverage | UMI | Amplification Technology |
|---|---|---|---|---|
| Tang | polyA | full length | No | PCR after polyA tailing |
| Quartz-seq | polyA | full length | No | |
| Quartz-seq2 | polyA | full length | Yes | |
| SUPeR-seq | polyA | full length | No | |
| MATQ-seq | polyA | full length | Yes | |
| SMART-seq | polyA | full length | No | Template-switching-based PCR |
| SMART-seq2 | polyA | full length | No | |
| SMART-seq3 | polyA | full length | Yes | |
| FLASH-seq | polyA | full length | Yes | |
| STRT-seq | polyA | 5′ tag | Yes | |
| STRT-seq-2i | polyA | 5′ tag | Yes | |
| SCRB-seq | polyA | 3′ tag | Yes | |
| Drop-seq | polyA | 3′ tag | Yes | |
| CEL-seq | polyA | 3′ tag | Yes | In vitro transcription(IVT) |
| CEL-seq2 | polyA | 3′ tag | Yes | |
| MARS-seq | polyA | 3′ tag | Yes | |
| MARS-seq2.0 | polyA | 3′ tag | Yes | |
| In Drops | polyA | 3′ tag | Yes |
| Applications | Category | Study | Clinical Significance | References |
|---|---|---|---|---|
| Heterogeneity | Heterogeneity within normal breast tissues | [ , ] | ||
| Heterogeneity within breast tumors | Offering insights into the refined classification and tailored therapies for breast cancer. | [ , , , ] | ||
| Heterogeneity between breast cancer malignant cells and reference normal epithelial cells | Revealing evolution mimicry during the specification of breast cancer subtype. | Revealing the origin of tumor cells and providing a foundation for accurate prognostic and therapeutic stratification of breast cancer. | [ ] | |
| Heterogeneity among breast cancer cell lines | Investigation of the functional relationship among different cell subtypes in breast cancer cell lines and how this interdependence contributes to tumor development. | Highlighting the systemic nature of cancer and task stratification of cell populations to maintain tumor hallmarks. | [ ] | |
| Heterogeneity in gene expression within each tumor | Revealing the phenotypes and biology underlying the genetic evolution and clinical behavior of TNBC. | Highlighting the connection between the functional heterogeneity of TNBC and genomic evolution, and revealing the biological principles that lead to the poor prognosis of TNBC. | [ ] | |
| TME | Tumor immune microenvironment(T cells, B cells, macrophages, NK cells) | [ , , , , , ] | ||
| Tumor interstitial microenvironment (CAFs) | [ , , , , ] | |||
| Therapy | Drug sensitivity | Predicting drug sensitivity | Guiding personalized drug treatment for patients. | [ ] |
| Predictive markers for NAT | Screening for biomarkers associated with the prognostic response to NAT. | Enabling the identification of subgroups of breast cancer patients who are likely to benefit from NAT. | [ , , ] | |
| Chemotherapy combined with immunotherapy | Analyses on the changes in the immune microenvironment and immune cell dynamics of breast cancer resulting from chemotherapy combined with immunotherapy. | Highlighting the role and concerns of specific immune cells in combined therapy, which could potentially provide important clues for individualized treatment. | [ , ] | |
| Drug resistance | Drug resistance of TNBC | [ , ] | ||
| Drug resistance of luminal breast cancer | [ , ] | |||
| Drug resistance of HER2-positive breast cancer | [ , ] | |||
| Drug resistance of non-inflammatory breast cancer | The role of combined application of MSA-2 and YM101 in immune therapy resistance of non-inflammatory tumors. | Providing a new treatment strategy for non-inflammatory tumors. | [ ] | |
| Metastasis | Lymph node metastasis in female breast cancer | [ , , , , , ] | ||
| Metastasis in male breast cancer | Metastatic characteristics of male breast cancer. | Providing a new perspective for the research and treatment of male breast cancer. | [ ] |
| The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
Share and Cite
Xiang, L.; Rao, J.; Yuan, J.; Xie, T.; Yan, H. Single-Cell RNA-Sequencing: Opening New Horizons for Breast Cancer Research. Int. J. Mol. Sci. 2024 , 25 , 9482. https://doi.org/10.3390/ijms25179482
Xiang L, Rao J, Yuan J, Xie T, Yan H. Single-Cell RNA-Sequencing: Opening New Horizons for Breast Cancer Research. International Journal of Molecular Sciences . 2024; 25(17):9482. https://doi.org/10.3390/ijms25179482
Xiang, Lingyan, Jie Rao, Jingping Yuan, Ting Xie, and Honglin Yan. 2024. "Single-Cell RNA-Sequencing: Opening New Horizons for Breast Cancer Research" International Journal of Molecular Sciences 25, no. 17: 9482. https://doi.org/10.3390/ijms25179482
Article Metrics
Article access statistics, further information, mdpi initiatives, follow mdpi.

Subscribe to receive issue release notifications and newsletters from MDPI journals
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 06 September 2024
GATA3 and markers of epithelial-mesenchymal transition predict long-term benefit from tamoxifen in ER-positive breast cancer
- Josefine Sandström 1 ,
- Jens Bomanson 1 ,
- Gizeh Pérez-Tenorio 1 ,
- Carolin Jönsson 1 ,
- Bo Nordenskjöld 1 ,
- Tommy Fornander 2 ,
- Linda S. Lindström ORCID: orcid.org/0000-0002-7722-7532 2 , 3 &
- Olle Stål ORCID: orcid.org/0000-0002-8290-0592 1
npj Breast Cancer volume 10 , Article number: 78 ( 2024 ) Cite this article
Metrics details
- Breast cancer
GATA binding protein 3 (GATA3) is essential for normal development of the mammary gland and associated with ER-positive breast cancer. Loss of GATA3 has been associated with epithelial-mesenchymal transition (EMT) in experimental studies. We investigated tumoral GATA3 in a cohort of postmenopausal patients with lymph-node negative breast cancer, randomized to adjuvant tamoxifen or control. Nuclear GATA3 expression was assessed with immunohistochemistry and GATA3 gene expression with Agilent microarrays. High GATA3 nuclear expression was associated with a lower rate of distant recurrence in ER-positive breast cancer (HR = 0.60, 95% CI 0.39–0.93). Low gene expression of GATA3 was associated with limited long-term benefit from adjuvant tamoxifen (interaction: p = 0.033). GATA3 gene expression was associated with the epithelial markers CDH1 (E-cadherin) and FOXA1, whereas negatively associated with several mesenchymal markers. Low expression of CDH1 was associated with marginal tamoxifen benefit (HR = 0.80 (0.43–1.49)), whereas patients with higher expression showed a significant benefit (HR = 0.33 (0.20–0.55), interaction: p = 0.029). In ER-positive breast cancer, diminished expression of GATA3 is associated with markers of EMT and poor long-term benefit from tamoxifen.
Introduction
Breast cancer commonly arises in luminal cells of the mammary gland expressing the estrogen receptor (ER). The expression of ER in the tumor is a cornerstone for the selection of adjuvant treatment. Patients with ER-positive breast cancer receiving endocrine therapy have initially a good prognosis, but studies with long-term follow-up have shown that there is a continuous risk of late relapse of the disease for these patients 1 , 2 , 3 . The endocrine treatment lasts for five or even ten years and there is a need for better prediction of overtreatment as well as of development of treatment resistance and late relapse.
The range of target genes regulated by ER is dependent on the phosphorylation of ER at different sites and cofactors that bind to DNA in the proximity of ER-binding sites 4 . GATA binding protein 3 (GATA3) and forkhead box A1 (FOXA1) are two proteins forming a strong transcriptional network together with ER. This network is required for correct development of the mammary gland 5 , 6 . ER-positive tumors often express GATA3, but a low GATA3 expression has been associated with worse prognosis 7 , 8 . However, whether GATA3 is an independent prognostic factor, and whether it predicts the benefit from tamoxifen, has yet not been settled. The importance of phosphorylated ER (pER) for the efficacy of tamoxifen has been previously studied by others and by us 9 , 10 , 11 , 12 . The receptor is phosphorylated at different sites by intracellular signaling molecules including MAP-kinase (pERser118), S6K1 (pERser167) and, PKA and PAK1 (pERser305) 11 , 12 , 13 , 14 .
Loss of functional GATA3 can arise due to mutations in the gene, which have been found in 10–15% of ER-positive breast cancer 15 . Mutations seldom silence the GATA3 gene- or protein expression but could alter its function. Without functional GATA3, or perhaps with too much of a function, the transcriptional landscape will change. Experimental studies suggest that loss of GATA3 expression is associated with epithelial-mesenchymal transition (EMT), which in turn might facilitate dissemination of tumor cells and the establishment of metastases 16 . To the best of our knowledge, the relationship between GATA3 and EMT factors in human breast tumors has not been thoroughly investigated.
The functionality of GATA3 can be disrupted by gene mutations or could be reduced by decreased gene expression or increased protein degradation. Here, we investigate the tumoral expression of GATA3 in a cohort of postmenopausal patients with lymph-node negative breast cancer, randomized to adjuvant tamoxifen or no systemic treatment. With gene expression data from the same cohort, we analyze several genes involved in EMT and cell adhesion in relation to GATA3 in ER-positive breast cancer. Furthermore, we examine the long-term prognosis and benefit from adjuvant tamoxifen in association to GATA3 and related factors.
Patient cohort
In a trial conducted by the Stockholm Breast Cancer Study Group, postmenopausal breast cancer patients with a negative lymph node status and tumor size not exceeding three cm were randomized to adjuvant tamoxifen, 40 mg daily for two years or no tamoxifen, the Stockholm tamoxifen trial STO-3 17 . The trial recruited patients regardless of ER status. Patient entry to the study was from November 1976 to May 1990. In 1983, tamoxifen-treated recurrence-free patients were randomized, if consenting, to three more years of tamoxifen or no further treatment with tamoxifen. The follow-up period lasted at most to 30 years with a median of 22.6 years of follow-up.
Figure 1 shows a flowchart of the study cohort. For 912 of the 1780 patients in the trial, formalin-fixed paraffin-embedded tumor tissue was available and used to construct tissue micro arrays (TMAs) with three tissue cores from each tumor. Previously, data for samples on the TMA were compared with data of the original cohort of 1780 patients 9 . The results showed no bias with respect to tumor size, ER status, or treatment arm. The STO‐3 trial was approved by the ethics committee at Karolinska Institutet (KI) in Stockholm, Sweden, and participants provided oral consent (KI 76–51). Further, the ethics committee at KI approved retrospective studies on archived tumor tissue for the present cohort, with the purpose to evaluate prognostic and treatment predicting factors (KI 97–451 with amendments 030201 and 2017 2066-32). Further need for patient consent was waived by the ethics committee.

Flowchart of included patients. Formalin-fixed paraffin-embedded (FFPE), Gene expression (GEX).
Biomarkers previously investigated in this cohort
Immunohistochemistry of ER, progesterone receptor (PgR) and HER2 was previously assessed 18 , 19 . Further, for ER phosphorylated at two different sites (pER), pERs167 and pERs305, data were available 9 , 10 . The antibodies used for ER, PgR and HER2 were, respectively, the CONFIRM™ mouse anti-ER antibody (clone 6F11) and the CONFIRM™ mouse anti-PR antibody (clone 16) from Ventana Medical Systems, and the DAKO AO0485 polyclonal rabbit antibody according to the guidelines provided by the manufacturer. The antibodies for pER were a rabbit polyclonal pERα ser305 primary antibody (Bethyl Laboratories, Montgomery, TX) and anti-pERs167 from Cell Signaling Technologies (Danvers, MA).
GATA3 protein staining
The GATA3 protein was investigated with immunohistochemistry on TMA sections. Several studies comparing antibody sensitivity revealed the L50-823 as the most sensitive GATA3 antibody compared to another commonly used antibody, HG3-31, reviewed in Kandalaft et al. 20 . The PT-link station was used for deparaffinization and antigen retrieval in a low-pH buffer (K800521-2, EnVision FLEX Target Retrieval Solution, DakoCytomation, Glostrup, Denmark), starting at 65 °C, gradually increasing and ending at 96 °C for 20 min and cooled down to 65 °C. Inactivation of endogenous peroxidase in 3% hydrogen peroxide in water was followed by blocking in serum-free protein block for 10 min (DPB-125, Spring Bioscience, Freemont, CA). TMA sections were incubated in a moisturized chamber at 4 °C during 24 h with the anti-GATA3 mouse monoclonal antibody diluted 1:500 (L50-823, Merck KGaA, Darmstadt, Germany). Secondary anti-mouse antibody (K4000, DakoCytomation Envision+ HRP system, Agilent Technologies, CA) was applied for 30 min at room temperature and protein staining was developed with 3,3’-diaminobenzidine chromogen and substrate buffer, dilution 1:50, for 8 min (K3467, DakoCytomation, Agilent Technologies, CA) and counterstained with hematoxylin for 1 min. All washing steps were in phosphate buffered saline including 0.5% bovine serum albumin. The tissue was dehydrated, and cover glass was mounted with Pertex (00871, Histolab, Askim, Sweden). Slides were visualized using the Aperio CS2 brightfield digital scanner at ×400 magnification and analyzed with the ImageScope software (Leica biosystems, Buffalo Grove, IL).
GATA3 protein grading
Of the 912 tumors, 749 were successfully stained and graded for GATA3 protein expression. Nuclear staining intensity was graded in three steps: negative (0), weak (1) and strong (2). Frequency of positive tumor nuclei were scored as follows; 0% (0), 1–10% (1), 11–50% (2), 51–89% (3) and ≥90% (4). For statistical analysis, the nuclear staining was divided in low and high, with a cut-off defining the group with high expression as strong nuclear staining in >50% of tumor cells. Primary grading was performed by two independent observers (J.S. and J.B.), blinded to clinical data, and secondary grading was performed jointly by the two observers to reach consensus.
Gene expression analysis
Messenger RNA was extracted from FFPE breast tumor tissue and 652 samples were available for microarray gene-expression analysis using custom-designed arrays, containing 32.1 K probes, detecting about 21.5 K unique genes (Agilent Technologies, CA) 21 . The Prediction Analysis of Microarray 50 (PAM50) intrinsic subtype analysis classifier was used as described by Parker et al. 22 .
We selected a number of genes encoding proteins known to interact with GATA3 or EMT. FOXA1 is a strong transcriptional partner of GATA3 and NOTCH3 might upregulate GATA3. Several factors related to EMT are known from the literature. Among them, we selected E-cadherin (CDH1), an epithelial marker frequently lost in EMT. On the other hand, N-cadherin (CDH2) and the intermediate filament protein vimentin (VIM) are expressed in mesenchymal cells. In addition, we selected alpha-smooth muscle actin (ACTA2) that is associated with the TGF-β pathway, which in turn can drive the EMT process. Finally, the EMT-activating transcription factors Snail (SNAI1) and Twist-related protein 1 (TWIST1) were included in the list of genes analyzed.
Expression levels of GATA3 and other genes were analyzed by tertiles (T1-T3) in the association analyses. In the survival analyses, the lowest tertile (T1) was used as cut-off for GATA3, CDH1 and FOXA1, as the low expression was expected to stand out from the group as more aggressive. For the EMT-related genes, the cut-off was set at the highest tertile (T3), as a high expression of these genes was expected to stand out from the remaining group as more aggressive. In the multiple regression analysis, the gene-expression levels were analyzed as continuous variables.
Statistical analyses were performed using Statistica 14 (TIBCO Software Inc.). For comparisons of GATA3 protein expression with other characteristics, the Pearson χ2 test was applied for 2 × 2 tables. For associations of GATA3 gene expression in three categories with other factors, the Spearman rank order correlation was applied. Distribution of GATA3 in the PAM50 molecular signatures was compared with the Kruskal–Wallis test. Multiple linear regression analysis was used to investigate how each mesenchymal marker could be predicted by GATA3, FOXA1 and NOTCH3. Distant recurrence-free interval (DRFI) time distributions were compared, and plots were drawn with the Kaplan–Meier method, visualizing time from randomization to first event of distant metastasis. Hazard ratios (HRs) of distant metastasis were estimated using the Cox proportional hazards model. Cox models were furthermore applied in multivariable analysis and in interaction analysis exploring the expression of genes as potential predictive factors for tamoxifen treatment benefit. A p -value of less than 0.05 was considered significant.
Overall, 70% of the tumors exhibited high nuclear GATA3 expression and GATA3 was strongly associated with ER status. Among ER-positive tumors, high nuclear and mRNA GATA3 expression, was seen in 84% and 41% of the cases, respectively. Corresponding numbers for ER-negative tumors were 19% and 5%, respectively ( p < 0.0001 for both). Accordingly, the PAM50 molecular subtypes differed in relation to GATA3, for both nuclear and mRNA expression, with Luminal A showing the highest levels and the Basal subtype the lowest (Fig. 2 ). Therefore, we restricted the analyses in the following to patients with ER-positive breast cancer and the tertiles for GATA3 were from now on based on this subgroup.

The fraction of tumors with high GATA3 protein expression ( a ) and gene expression levels ( b ) in relation to the PAM50 subtypes. Kruskal–Wallis H-test, p < 0.0001 for both comparisons.
Associations of GATA3 with clinicopathological variables and pER
Nuclear GATA3 was more frequently expressed at high levels in small and PgR-positive tumors (Table 1 ). HER2 positivity was significantly associated with low GATA3 gene expression levels and a similar trend was seen for GATA3 protein expression (Tables 1 and 2 ). Both protein and gene expression levels of GATA3 were associated with ER phosphorylated at serine-305 (pERs305), whereas the association of GATA3 with pERs167 was less clear.
There was a significant correlation between GATA3 mRNA and protein levels (Table 1 ), but this relationship showed differences dependent on pER. For tumors with a positive status of pERs167, nuclear GATA3 was frequently highly expressed also at low GATA3 mRNA levels, resulting in no correlation between gene and protein expression levels.
On the other hand, the subgroup of pERs167- negative tumors showed a significant gene to protein correlation (Fig. 3 ). In contrast, the status of pERs305 did not markedly affect the correlation between gene and protein expression levels.

The relationship between GATA3 gene and protein expression by the status of pERs167 ( a ) and pERs305 ( b ). The p -values refer to Spearman rank order correlation and error bars refer to standard error of the mean. Tertile (T).
Associations of GATA3 with epithelial and mesenchymal biomarkers
Based on previous experimental research showing that the loss of GATA3 may contribute to EMT, we next analyzed the relationship between GATA3 and the expression of several genes associated with this process. Comparing gene-expression levels, GATA3 was negatively correlated with all the mesenchymal biomarkers investigated, including ACTA2, CDH2, SNAI1, TWIST1 and VIM (Table 3 ). For GATA3 protein levels, the same was true for three of the biomarkers. Furthermore, the gene expression of GATA3 was positively associated with CDH1, FOXA1, and NOTCH3.
Since the transcription factors GATA3 and FOXA1 have been suggested to inhibit EMT, and NOTCH3 could be a contributing factor, we performed multiple regression analysis to investigate these factors as independent predictors of CDH1 and the mesenchymal markers, respectively (Table 4 ). GATA3 turned out to be the variable that most consistently correlated with the markers investigated.
Distant recurrence-free interval in relation to GATA3
Patients with high tumoral nuclear GATA3 expression had a longer DRFI than those with low GATA3 levels (HR = 0.60, 95% CI 0.39–0.93, p = 0.023, Fig. 4a ). When adjusting for treatment and other tumor characteristics, including tumor size, PgR and HER2, the statistical significance for GATA3 was lost (HR = 0.81, 95% CI 0.50–1.31, p = 0.39), whereas tumor size (>20 mm vs ≤20 mm; HR = 2.16, 95% CI 1.39–3.36, p = 0.00067) and tamoxifen (HR = 0.45, 95% CI 0.30–0.67, p < 0.0001) were significant. We did not find a significant association of GATA3 gene expression levels with DRFI (T2-T3 vs T1; HR = 1.06, 95% CI 0.71–1.57, p = 0.78, Fig. 4b ).

Distant recurrence-free interval (DRFI) in relation to GATA 3 nuclear expression ( a ) and GATA3 gene expression ( b ). HR Hazard ratio, CI Confidence interval.
The benefit from adjuvant tamoxifen in relation to GATA3 and related factors
Given that GATA3 and ER interact, the benefit from tamoxifen could potentially be dependent on expression levels of GATA3. Whereas patients with ER-positive tumors with intermediate to high GATA3 mRNA levels showed significant benefit from tamoxifen (HR = 0.39 (0.24–0.64), p = 0.00014), those with levels in the bottom tertile did not as evidently benefit (HR = 0.61 (0.31–1.17), p = 0.14) (Fig. 5 ). The difference was more pronounced when considering the long-term prognosis. For patients still alive and without a distant recurrence after five years, there was no further benefit from tamoxifen in the group with low GATA3 (HR = 1.10 (0.46–2.61), p = 0.83), in contrast to the group with higher levels (HR = 0.35 (0.19–0.64), p = 0.00064). A test for interaction between GATA3 and tamoxifen for this period was significant ( p = 0.033). The efficacy of tamoxifen did not significantly differ for patients with low or high tumoral nuclear GATA3 protein expression (HR = 0.53 (0.23–1.23) and HR = 0.52 (0.34–0.79), respectively), however the number of patients in the former group was small.

DRFI for the tamoxifen and control groups in patients with low ( a ) and medium/high ( b ) GATA3 levels. DRFI for the tamoxifen and control groups, in patients alive and free of distant recurrence after five years, with low ( c ) and medium/high ( d ) GATA3. Hazard ratio (HR), tamoxifen (TAM), tertile (T).
The GATA3-associated genes CDH1 and FOXA1 in addition tended to predict the efficacy of tamoxifen. Patients with CDH1 levels in the bottom tertile experienced no evident benefit from tamoxifen (HR = 0.80 (0.43–1.49), p = 0.49), whereas those with higher levels did (HR = 0.33 (0.20–0.55), p = 0.000021), and the interaction was significant ( p = 0.029, Fig. 6a, b ). Although a similar interaction between FOXA1 and tamoxifen did not reach statistical significance ( p = 0.22), the benefit from the treatment was more evident in the group with higher levels as compared with the group with low levels (HR = 0.41 (0.26–0.63), p = 0.000075) and (HR = 0.71 (0.32–1.58), p = 0.40), respectively, (Fig. 6c, d ). Moreover, the EMT biomarkers CDH2 and VIM tended to predict less tamoxifen benefit when expressed at higher levels as compared to low levels (Fig. 6E–H ). For the remaining EMT markers, the benefit from tamoxifen was similar comparing groups with low/intermediate versus high levels (tests for interaction, all p > 0.7).

DRFI for the tamoxifen and controls groups in patients with low ( a ) and medium/high tumor CDH1 ( b ), in patients with low ( c ) and medium/high tumor FOXA1 ( d ), in patients with low ( e ) and medium/high tumor CDH2 ( f ), in patients with low ( g ) and medium/high tumor VIM ( h ). Hazard ratio (HR), tamoxifen (TAM), tertile (T).
GATA3, known as a marker used to identify mammary or urothelial origin of metastases from unknown primary tumors, is widely expressed in breast tumors and has been suggested as a potential prognostic and/or treatment predictive biomarker 23 , 24 . A quantitative decrease or functional loss of GATA3 seems to interfere with the characteristics of the tumor 25 . In line with previously published data, we see a distinct nuclear expression of GATA3 in more than 80% of ER-positive tumors as compared to in only about 20% of the ER-negative tumors 7 , 26 , 27 . GATA3 positivity was largely associated with a PAM50 Luminal subtype, supporting its functional role as co-regulator of the ER and its association with differentiation.
The ER protein is phosphorylated at distinct sites when regulated. The tight connection between ER and GATA3 suggests that the two proteins may regulate each other. We found a significant association between pERs305 and high GATA3 expression. We suggest that this might be related to PKA, which, besides its involvement in ERs305 phosphorylation 11 , has been shown to interact with GATA3 28 . Overall, there was a strong association of GATA3 gene expression with GATA3 protein expression. Interestingly, in tumors phosphorylated at ERs167, GATA3 gene- and protein expression did not correlate. Hypothetically, a stabilization of the GATA3 protein could be affected by intracellular signaling proteins associated with ER phosphorylation, supported by the finding that MAPK controls GATA3 protein stability by a post-transcriptional mechanism 29 . Furthermore, S6K1, that phosphorylates ER at serine 167, interacts with ER in a positive feedback loop also involving GATA3 30 .
We found low GATA3 protein levels to be significantly associated with increased risk of distant recurrence when compared to high GATA3 protein levels in the group of patients with ER-positive tumors. Mehra et al. reported already in 2005 that detection of GATA3 with immunohistochemistry could predict outcome of breast cancer, also when adjusting for other prognostic factors 8 . Several studies have found similar results 7 , 27 , 31 , although the independent prognostic value was absent in one of the studies when the analysis was restricted to ER-positive breast cancer 31 , similar to our results. Moreover, GATA3 was not independently prognostic in a huge study comprising more than 3000 patients 32 . We did not see a prognostic value of GATA3 gene-expression levels in the present cohort. This contrasts with some other gene-expression studies with microarray data, reviewed by Fang et al, showing that GATA3 is prognostic 33 . Taken together, GATA3 appears as a favorable prognostic factor, but the question of its importance as an independent factor needs further elucidation, considering the choice of cut-off for positivity as well as the influence of breast cancer therapy.
GATA3 is highly correlated with ER and thus a potential predictive marker of benefit from hormonal therapy. Using a breast cancer model with parental and tamoxifen-resistant MCF7 cells, endocrine resistance was associated with downregulation of luminal/epithelial differentiation markers and upregulation of basal/mesenchymal invasive markers 34 . One important factor for the transcriptional landscape was GATA3. Further studies have indicated that GATA3 counteracts EMT. The protein complex GATA3/G9A/MTA3 represses ZEB2, and other genes involved in EMT, leading to suppression of metastasis from human breast cancer cells in mice. In turn, ZEB2 repressed the expression of G9A and MTA3 35 . Moreover, ectopic expression of GATA3 in GATA3-negative triple-negative breast cancer cells led to increased CDH1 expression and decreased expression of some mesenchymal markers 16 . A similar transcriptional change could also be related to mutations of GATA3 36 . Here, the results give support for the experimental findings of an inverse relationship between GATA3 and EMT in a large series of ER-positive tumors. GATA3 expression correlated with high expression of E-cadherin and FOXA1 and low expressions of all five mesenchymal markers investigated.
Besides a prognostic value, one could ask whether GATA3 is a treatment predictive factor. In a minor series of 28 ER-positive cases of breast cancer, lack of GATA3 expression was associated with unresponsiveness to hormonal therapy 37 . Furthermore, GATA3 mRNA expression was associated with longer progression-free survival in patients with ER-positive breast cancer treated with first-line tamoxifen for recurrent disease 38 . To the best of our knowledge, there are no previous reports on the relationship of GATA3 with the efficacy of adjuvant endocrine therapy based on a randomized trial. In the present study, we were able to show that a substantial benefit from tamoxifen was restricted to patients with intermediate/high GATA3 mRNA expression, most evident when focusing on late relapse. We found similar patterns for FOXA1 and E-cadherin, both of which are closely related to GATA3, whereas CDH2 (N-cadherin) and VIM (vimentin) tended to show opposite associations. When the transcriptional landscape of ER is altered upon loss of GATA3, one might speculate that the anti-tumoral effects of tamoxifen is diminished.
One strength with this study is that it is based on a randomized clinical trial and long-term follow-up. The study limitations include that the cohort is confined to postmenopausal patients with lymph-node negative disease, and it is not known if the results related to tamoxifen benefit can be translated to the use of aromatase inhibitors. Another limitation is that we lack data on GATA3 mutations for this cohort. GATA3 is the third most mutated gene in luminal breast cancer. In part, the results of the present study might be applicable to tumors with GATA3 mutation as such mutations affect gene transcription patterns and EMT 36 , 39 , 40 .
In conclusion, GATA3 expression is associated with ER-positive breast cancer and particularly with the Luminal A subtype. Diminished expression of GATA3 in ER-positive tumors is associated with changes of gene expression resembling EMT. Both such changes and GATA3 expression itself were related to the efficacy of adjuvant tamoxifen therapy.
Data availability
Restrictions apply to the availability of these data according to GDPR. Data were obtained from the STO Trialist Group and are available from the authors with the permission from the STO Trialist Group.
Pan, H. et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N. Engl. J. Med. 377 , 1836–1846 (2017).
Article PubMed PubMed Central Google Scholar
Dar, H. et al. Assessment of 25-Year Survival of Women With Estrogen Receptor-Positive/ERBB2-Negative Breast Cancer Treated With and Without Tamoxifen Therapy: A Secondary Analysis of Data From the Stockholm Tamoxifen Randomized Clinical Trial. JAMA Netw. Open 4 , e2114904 (2021).
Johansson, A. et al. Twenty-Year Benefit From Adjuvant Goserelin and Tamoxifen in Premenopausal Patients With Breast Cancer in a Controlled Randomized Clinical Trial. J. Clin. Oncol. 40 , 4071–4082 (2022).
Article CAS PubMed PubMed Central Google Scholar
Faus, H. & Haendler, B. Post-translational modifications of steroid receptors. Biomed. Pharmacother. 60 , 520–528 (2006).
Article CAS PubMed Google Scholar
Kouros-Mehr, H., Slorach, E. M., Sternlicht, M. D. & Werb, Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 127 , 1041–1055 (2006).
Chou, J., Provot, S. & Werb, Z. GATA3 in development and cancer differentiation: cells GATA have it! J. Cell Physiol. 222 , 42–49 (2010).
Yoon, N. K. et al. Higher levels of GATA3 predict better survival in women with breast cancer. Hum. Pathol. 41 , 1794–1801 (2010).
Mehra, R. et al. Identification of GATA3 as a breast cancer prognostic marker by global gene expression meta-analysis. Cancer Res. 65 , 11259–11264 (2005).
Bostner, J., Skoog, L., Fornander, T., Nordenskjold, B. & Stal, O. Estrogen receptor-alpha phosphorylation at serine 305, nuclear p21-activated kinase 1 expression, and response to tamoxifen in postmenopausal breast cancer. Clin. Cancer Res. 16 , 1624–1633 (2010).
Bostner, J. et al. Activation of Akt, mTOR, and the estrogen receptor as a signature to predict tamoxifen treatment benefit. Breast Cancer Res. Treat. 137 , 397–406 (2013).
Kok, M. et al. PKA-induced phosphorylation of ERalpha at serine 305 and high PAK1 levels is associated with sensitivity to tamoxifen in ER-positive breast cancer. Breast Cancer Res. Treat. 125 , 1–12 (2011).
Rayala, S. K., Molli, P. R. & Kumar, R. Nuclear p21-activated kinase 1 in breast cancer packs off tamoxifen sensitivity. Cancer Res. 66 , 5985–5988 (2006).
Kato, S. et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science 270 , 1491–1494 (1995).
Yamnik, R. L. et al. S6 kinase 1 regulates estrogen receptor alpha in control of breast cancer cell proliferation. J. Biol. Chem. 284 , 6361–6369 (2009).
Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature 490 , 61–70 (2012).
Article Google Scholar
Yan, W., Cao, Q. J., Arenas, R. B., Bentley, B. & Shao, R. GATA3 inhibits breast cancer metastasis through the reversal of epithelial-mesenchymal transition. J. Biol. Chem. 285 , 14042–14051 (2010).
Rutqvist, L. E., Johansson, H. & Stockholm Breast Cancer Study, G. Long-term follow-up of the randomized Stockholm trial on adjuvant tamoxifen among postmenopausal patients with early stage breast cancer. Acta Oncol. 46 , 133–145 (2007).
Khoshnoud, M. R. et al. Immunohistochemistry compared to cytosol assays for determination of estrogen receptor and prediction of the long-term effect of adjuvant tamoxifen. Breast Cancer Res. Treat. 126 , 421–430 (2011).
Jerevall, P. L. et al. Predictive relevance of HOXB13 protein expression for tamoxifen benefit in breast cancer. Breast Cancer Res. 12 , R53 (2010).
Kandalaft, P. L., Simon, R. A., Isacson, C. & Gown, A. M. Comparative Sensitivities and Specificities of Antibodies to Breast Markers GCDFP-15, Mammaglobin A, and Different Clones of Antibodies to GATA-3: A Study of 338 Tumors Using Whole Sections. Appl. Immunohistochem. Mol. Morphol. 24 , 609–614 (2016).
Esserman, L. J. et al. Use of Molecular Tools to Identify Patients With Indolent Breast Cancers With Ultralow Risk Over 2 Decades. JAMA Oncol. 3 , 1503–1510, (2017).
Article PubMed Google Scholar
Parker, J. S. et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27 , 1160–1167 (2009).
Khazaeli Najafabadi, M. et al. Role of GATA3 in tumor diagnosis: A review. Pathol. Res. Pr. 226 , 153611 (2021).
Article CAS Google Scholar
Bonacho, T., Rodrigues, F. & Liberal, J. Immunohistochemistry for diagnosis and prognosis of breast cancer: a review. Biotech. Histochem. 95 , 71–91 (2020).
Kouros-Mehr, H. et al. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell 13 , 141–152 (2008).
Gulbahce, H. E. et al. Significance of GATA-3 expression in outcomes of patients with breast cancer who received systemic chemotherapy and/or hormonal therapy and clinicopathologic features of GATA-3-positive tumors. Hum. Pathol. 44 , 2427–2431 (2013).
Querzoli, P. et al. GATA3 as an Adjunct Prognostic Factor in Breast Cancer Patients with Less Aggressive Disease: A Study with a Review of the Literature. Diagnostics 11 , https://doi.org/10.3390/diagnostics11040604 (2021).
Bouchard, M. F., Taniguchi, H. & Viger, R. S. Protein kinase A-dependent synergism between GATA factors and the nuclear receptor, liver receptor homolog-1, regulates human aromatase (CYP19) PII promoter activity in breast cancer cells. Endocrinology 146 , 4905–4916 (2005).
Yamashita, M. et al. Ras-ERK MAPK cascade regulates GATA3 stability and Th2 differentiation through ubiquitin-proteasome pathway. J. Biol. Chem. 280 , 29409–29419 (2005).
Maruani, D. M. et al. Estrogenic regulation of S6K1 expression creates a positive regulatory loop in control of breast cancer cell proliferation. Oncogene 31 , 5073–5080 (2012).
Jacquemier, J. et al. Association of GATA3, P53, Ki67 status and vascular peritumoral invasion are strongly prognostic in luminal breast cancer. Breast Cancer Res. 11 , R23 (2009).
Voduc, D., Cheang, M. & Nielsen, T. GATA-3 expression in breast cancer has a strong association with estrogen receptor but lacks independent prognostic value. Cancer Epidemiol. Biomark. Prev. 17 , 365–373 (2008).
Fang, S. H., Chen, Y. & Weigel, R. J. GATA-3 as a marker of hormone response in breast cancer. J. Surg. Res. 157 , 290–295 (2009).
Bi, M. et al. Enhancer reprogramming driven by high-order assemblies of transcription factors promotes phenotypic plasticity and breast cancer endocrine resistance. Nat. Cell Biol. 22 , 701–715 (2020).
Si, W. et al. Dysfunction of the Reciprocal Feedback Loop between GATA3- and ZEB2-Nucleated Repression Programs Contributes to Breast Cancer Metastasis. Cancer Cell 27 , 822–836 (2015).
Takaku, M., Grimm, S. A., De Kumar, B., Bennett, B. D. & Wade, P. A. Cancer-specific mutation of GATA3 disrupts the transcriptional regulatory network governed by Estrogen Receptor alpha, FOXA1 and GATA3. Nucleic Acids Res. 48 , 4756–4768 (2020).
Parikh, P., Palazzo, J. P., Rose, L. J., Daskalakis, C. & Weigel, R. J. GATA-3 expression as a predictor of hormone response in breast cancer. J. Am. Coll. Surg. 200 , 705–710 (2005).
Liu, J. et al. GATA3 mRNA expression, but not mutation, associates with longer progression-free survival in ER-positive breast cancer patients treated with first-line tamoxifen for recurrent disease. Cancer Lett. 376 , 104–109 (2016).
Saotome, M., Poduval, D. B., Nair, R., Cooper, M. & Takaku, M. GATA3 Truncation Mutants Alter EMT Related Gene Expression via Partial Motif Recognition in Luminal Breast Cancer Cells. Front. Genet 13 , 820532 (2022).
Cohen, H. et al. Shift in GATA3 functions, and GATA3 mutations, control progression and clinical presentation in breast cancer. Breast Cancer Res. 16 , 464 (2014).
Download references
Acknowledgements
This study was funded by the Swedish Cancer Society (Cancerfonden, grant No. 190268 to O.S. and grant No. 222081 and 220552SIA to L.S.L., ALF medicine (grant number Lio-795201, O.S. and grant number FoUI-974882 to L.S.L.), Onkologiska Klinikernas i Linköpings forskningsfond, 2019 to J.S., Swedish Research Council (Vetenskapsrådet, grant number 2020-02466 to L.S.L.). The funders played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Open access funding provided by Linköping University.
Author information
Authors and affiliations.
Department of Biomedical and Clinical Sciences and Department of Oncology, 581 83 Linköping University, Linköping, Sweden
Josefine Sandström, Jens Bomanson, Gizeh Pérez-Tenorio, Carolin Jönsson, Bo Nordenskjöld & Olle Stål
Department of Oncology and Pathology, Karolinska Institute and University Hospital, Stockholm, Sweden
Tommy Fornander & Linda S. Lindström
Breast Center, Karolinska Comprehensive Cancer Center, Karolinska University Hospital, Stockholm, Sweden
Linda S. Lindström
You can also search for this author in PubMed Google Scholar
Contributions
Conception and design: J.S., O.S. Data acquisition: J.S., J.B., B.N., T.F., L.S.L. Formal analysis: J.S., J.B., O.S. Data interpretation: all authors. Funding acquisition: J.S., O.S., L.S.L. Writing—original draft: J.S., O.S. Writing—review & editing: all authors. The final version was approved by all authors.
Corresponding author
Correspondence to Olle Stål .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Sandström, J., Bomanson, J., Pérez-Tenorio, G. et al. GATA3 and markers of epithelial-mesenchymal transition predict long-term benefit from tamoxifen in ER-positive breast cancer. npj Breast Cancer 10 , 78 (2024). https://doi.org/10.1038/s41523-024-00688-6
Download citation
Received : 17 April 2024
Accepted : 29 August 2024
Published : 06 September 2024
DOI : https://doi.org/10.1038/s41523-024-00688-6
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Cancer newsletter — what matters in cancer research, free to your inbox weekly.
Advances in Breast Cancer Research
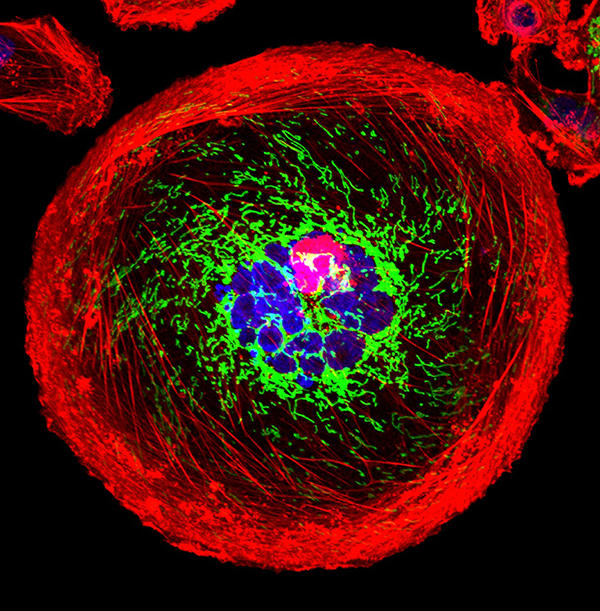
A polyploid giant cancer cell (PGCC) from triple-negative breast cancer.
NCI-funded researchers are working to advance our understanding of how to prevent, detect, and treat breast cancer. They are also looking at how to address disparities and improve quality of life for survivors of the disease.
This page highlights some of what's new in the latest research for breast cancer, including new clinical advances that may soon translate into improved care, NCI-supported programs that are fueling progress, and research findings from recent studies.
Early Detection of Breast Cancer
Breast cancer is one of a few cancers for which an effective screening test, mammography , is available. MRI ( magnetic resonance imaging ) and ultrasound are also used to detect breast cancer, but not as routine screening tools for people with average risk.
Ongoing studies are looking at ways to enhance current breast cancer screening options. Technological advances in imaging are creating new opportunities for improvements in both screening and early detection.
One technology advance is 3-D mammography , also called breast tomosynthesis . This procedure takes images from different angles around the breast and builds them into a 3-D-like image. Although this technology is increasingly available in the clinic, it isn’t known whether it is better than standard 2-D mammography , for detecting cancer at a less advanced stage.
NCI is funding a large-scale randomized breast screening trial, the Tomosynthesis Mammographic Imaging Screening Trial (TMIST) , to compare the number of advanced cancers detected in women screened for 5 years with 3-D mammography with the number detected in women screened with 2-D mammography.
Two concerns in breast cancer screening, as in all cancer screening, are:
- the potential for diagnosing tumors that would not have become life-threatening ( overdiagnosis )
- the possibility of receiving false-positive test results, and the anxiety that comes with follow-up tests or procedures
As cancer treatment is becoming more individualized, researchers are looking at ways to personalize breast cancer screening. They are studying screening methods that are appropriate for each woman’s level of risk and limit the possibility of overdiagnosis.
For example, the Women Informed to Screen Depending on Measures of Risk (WISDOM) study aims to determine if risk-based screening—that is, screening at intervals that are based on each woman’s risk as determined by her genetic makeup, family history , and other risk factors—is as safe, effective, and accepted as standard annual screening mammography.
WISDOM is also making a focused effort to enroll Black women in the trial. Past studies tended to contain a majority of White women and therefore, there is less data on how screening can benefit Black women. Researchers are taking a number of steps to include as many Black women as possible in the study while also increasing the diversity of all women enrolled.
Breast Cancer Treatment
The mainstays of breast cancer treatment are surgery , radiation , chemotherapy , hormone therapy , and targeted therapy . But scientists continue to study novel treatments and drugs, along with new combinations of existing treatments.
It is now known that breast cancer can be divided into subtypes based on whether they:
- are hormone receptor (HR) positive which means they express estrogen and/or progesterone receptors ( ER , PR )
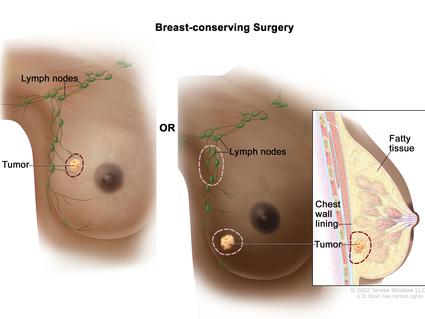
Shortening Radiation Therapy for Some with Early Breast Cancer
A condensed course was as effective and safe as the standard course for women with higher-risk early-stage breast cancer who had a lumpectomy.
As we learn more about the subtypes of breast cancer and their behavior, we can use this information to guide treatment decisions. For example:
- The NCI-sponsored TAILORx clinical trial. The study, which included patients with ER-positive, lymph node-negative breast cancer, found that a test that looks at the expression of certain genes can predict which women can safely avoid chemotherapy.
- The RxPONDER trial found that the same gene expression test can also be used to determine treatment options in women with more advanced breast cancer. The study found that some postmenopausal women with HR positive, HER-2 negative breast cancer that has spread to several lymph nodes and has a low risk of recurrence do not benefit from chemotherapy when added to their hormone therapy.
- The OFSET trial is comparing the addition of chemotherapy to usual treatment ( ovarian function suppression plus hormone therapy) to usual treatment alone in treating premenopausal estrogen receptor (ER)-positive/HER2-negative breast cancer patients who are at high risk of their cancer returning. This will help determine whether or not adding chemotherapy helps prevent the cancer from returning.
Genomic analyses, such as those carried out through The Cancer Genome Atlas (TCGA) , have provided more insights into the molecular diversity of breast cancer and eventually could help identify even more breast cancer subtypes. That knowledge, in turn, may lead to the development of therapies that target the genetic alterations that drive those cancer subtypes.
HR-Positive Breast Cancer Treatment
Hormone therapies have been a mainstay of treatment for HR-positive cancer. However, there is a new focus on adding targeted therapies to hormone therapy for advanced or metastatic HR-positive cancers. These treatments could prolong the time until chemotherapy is needed and ideally, extend survival. Approved drugs include:

Drug Combo Effective for Metastatic Breast Cancer in Younger Women
Ribociclib plus hormone therapy were superior to standard chemotherapy combos in a recent trial.
- Palbociclib (Ibrance) , ribociclib (Kisqali) , and everolimus (Afinitor) have all been approved by the FDA for use with hormone therapy for treatment of advanced or metastatic breast cancer. Ribociclib has been shown to increase the survival of patients with metastatic breast cancer . It has also shown to slow the growth of metastatic cancer in younger women when combined with hormone therapy.
- Elacestrant (Orserdu) is approved for HR-positive and HER2-negative breast cancer that has a mutation in the ESR1 gene, and has spread. It is used in postmenopausal women and in men whose cancer has gotten worse after at least one type of hormone therapy.
- Abemaciclib (Verzenio) can be used with or after hormone therapy to treat advanced or metastatic HR-positive, HER2-negative breast cancer. In October 2021, the Food and Drug Administration ( FDA ) approved abemaciclib in combination with hormone therapy to treat some people who have had surgery for early-stage HR-positive, HER2-negative breast cancer.
- Alpelisib (Piqray) is approved to be used in combination with hormone therapy to treat advanced or metastatic HR-positive, HER2-negative breast cancers that have a mutation in the PIK3CA gene .
- Sacituzumab govitecan-hziy (Trodelvy) is used for HR-positive and HER2-negative breast cancer that has spread or can't be removed with surgery. It is used in those who have received hormone therapy and at least two previous treatments. It has shown to extend the amount of time that the disease doesn't get worse ( progression-free survival ) and also shown to improve overall survival .
HER2-Positive Breast Cancer Treatment
The FDA has approved a number of targeted therapies to treat HER2-positive breast cancer , including:
- Trastuzumab (Herceptin) has been approved to be used to prevent a relapse in patients with early-stage HER2-positive breast cancer.
- Pertuzumab (Perjeta) is used to treat metastatic HER2-positive breast cancer, and also both before surgery ( neoadjuvant ) and after surgery ( adjuvant therapy ).
- Trastuzumab and pertuzumab together can be used in combination with chemotherapy to prevent relapse in people with early-stage HER2-positive breast cancer. Both are also used together in metastatic disease, where they delay progression and improve overall survival.
- Trastuzumab deruxtecan (Enhertu) is approved for patients with advanced or metastatic HER2-positive breast cancer who have previously received a HER2-targeted treatment. A 2021 clinical trial showed that the drug lengthened the time that people with metastatic HER2-positive breast cancer lived without their cancer progressing. The trial also showed that it was better at shrinking tumors than another targeted drug, trastuzumab emtansine (Kadcyla).
- Tucatinib (Tukysa) is approved to be used in combination with trastuzumab and capecitabine (Xeloda) for HER2-positive breast cancer that cannot be removed with surgery or is metastatic. Tucatinib is able to cross the blood–brain barrier, which makes it especially useful for HER2-positive metastatic breast cancer, which tends to spread to the brain.
- Lapatinib (Tykerb) has been approved for treatment of some patients with HER2-positive advanced or metastatic breast cancer, together with capecitabine or letrozole.
- Neratinib Maleate (Nerlynx) can be used in patients with early-stage HER2-positive breast cancer and can also be used together with capecitabine (Xeloda) in some patients with advanced or metastatic disease.
- Ado-trastuzumab emtansine (Kadcyla) is approved to treat patients with metastatic HER2-positive breast cancer who have previously received trastuzumab and a taxane . It's also used in some patients with early-stage HER2-positive breast cancer who have completed therapy before surgery ( neoadjuvant ) and have residual disease at the time of surgery.
HER2-Low Breast Cancer
A newly defined subtype, HER2-low, accounts for more than half of all metastatic breast cancers. HER2-low tumors are defined as those whose cells contain lower levels of the HER2 protein on their surface. Such tumors have traditionally been classified as HER2-negative because they did not respond to drugs that target HER2.
However, in a clinical trial, trastuzumab deruxtecan (Enhertu) improved the survival of patients with HER2-low breast cancer compared with chemotherapy , and the drug is approved for use in such patients.

Immunotherapy Improves Survival in Triple-Negative Breast Cancer
For patients whose tumors had high PD-L1 levels, pembrolizumab with chemo helped them live longer.
Triple-Negative Breast Cancer Treatment
Triple-negative breast cancers (TNBC) are the hardest to treat because they lack both hormone receptors and HER2 overexpression , so they do not respond to therapies directed at these targets. Therefore, chemotherapy is the mainstay for treatment of TNBC. However, new treatments are starting to become available. These include:
- Sacituzumab govitecan-hziy (Trodelvy) is approved to treat patients with TNBC that has spread to other parts of the body . Patients must have received at least two prior therapies before receiving the drug.
- Pembrolizumab (Keytruda) is an immunotherapy drug that is approved to be used in combination with chemotherapy for patients with locally advanced or metastatic TNBC that has the PD-L1 protein. It may also be used before surgery (called neoadjuvant ) for patients with early-stage TNBC, regardless of their PD-L1 status.
- PARP inhibitors, which include olaparib (Lynparza) and talazoparib (Talzenna) , are approved to treat metastatic HER2-negative or triple-negative breast cancers in patients who have inherited a harmful BRCA gene mutation. Olaparib is also approved for use in certain patients with early-stage HER2-negative or triple-negative breast cancer.
- Drugs that block the androgen receptors or prevent androgen production are being tested in a subset of TNBC that express the androgen receptor.
For a complete list of drugs for breast cancer, see Drugs Approved for Breast Cancer .

NCI-Supported Breast Cancer Research Programs
Many NCI-funded researchers working at the NIH campus, as well as across the United States and world, are seeking ways to address breast cancer more effectively. Some research is basic, exploring questions as diverse as the biological underpinnings of cancer and the social factors that affect cancer risk. And some are more clinical, seeking to translate this basic information into improving patient outcomes. The programs listed below are a small sampling of NCI’s research efforts in breast cancer.
TMIST is a randomized breast screening trial that compares two Food and Drug Administration (FDA)-approved types of digital mammography, standard digital mammography (2-D) with a newer technology called tomosynthesis mammography (3-D).
The Breast Specialized Programs of Research Excellence (Breast SPOREs) are designed to quickly move basic scientific findings into clinical settings. The Breast SPOREs support the development of new therapies and technologies, and studies to better understand tumor resistance, diagnosis, prognosis, screening, prevention, and treatment of breast cancer.
The NCI Cancer Intervention and Surveillance Modeling Network (CISNET) focuses on using modeling to improve our understanding of how prevention, early detection, screening, and treatment affect breast cancer outcomes.
The Confluence Project , from NCI's Division of Cancer Epidemiology and Genetics (DCEG) , is developing a research resource that includes data from thousands of breast cancer patients and controls of different races and ethnicities. This resource will be used to identify genes that are associated with breast cancer risk, prognosis, subtypes, response to treatment, and second breast cancers. (DCEG conducts other breast cancer research as well.)
The Black Women’s Health Study (BWHS) Breast Cancer Risk Calculator allows health professionals to estimate a woman’s risk of developing invasive breast cancer over the next 5 years. With the NCI-funded effort, researchers developed a tool to estimate the risk of breast cancer in US Black women. The team that developed the tool hopes it will help guide more personalized decisions on when Black women—especially younger women—should begin breast cancer screening.
The goal of the Breast Cancer Surveillance Consortium (BCSC) , an NCI-funded program launched in 1994, is to enhance the understanding of breast cancer screening practices in the United States and their impact on the breast cancer's stage at diagnosis, survival rates, and mortality.
There are ongoing programs at NCI that support prevention and early detection research in different cancers, including breast cancer. Examples include:
- The Cancer Biomarkers Research Group , which promotes research in cancer biomarkers and manages the Early Detection Research Network (EDRN) . EDRN is a network of NCI-funded institutions that are collaborating to discover and validate early detection biomarkers. Within the EDRN, the Breast and Gynecologic Cancers Collaborative Group conducts research on breast and ovarian cancers.
- NCI's Division of Cancer Prevention houses the Breast and Gynecologic Cancer Research Group which conducts and fosters the development of research on the prevention and early detection of breast and gynecologic cancers.
Breast Cancer Survivorship Research
NCI’s Office of Cancer Survivorship, part of the Division of Cancer Control and Population Sciences (DCCPS), supports research projects throughout the country that study many issues related to breast cancer survivorship. Examples of studies funded include the impact of cancer and its treatment on physical functioning, emotional well-being, cognitive impairment , sleep disturbances, and cardiovascular health. Other studies focus on financial impacts, the effects on caregivers, models of care for survivors, and issues such as racial disparities and communication.
Breast Cancer Clinical Trials
NCI funds and oversees both early- and late-phase clinical trials to develop new treatments and improve patient care. Trials are available for breast cancer prevention , screening , and treatment .
Breast Cancer Research Results
The following are some of our latest news articles on breast cancer research and study updates:
- How Breast Cancer Risk Assessment Tools Work
- Can Some People with Breast Cancer Safely Skip Lymph Node Radiation?
- Study Adds to Debate about Mammography in Older Women
- Pausing Long-Term Breast Cancer Therapy to Become Pregnant Appears to Be Safe
- A Safer, Better Treatment Option for Some Younger Women with Breast Cancer
- Shorter Course of Radiation Is Effective, Safe for Some with Early-Stage Breast Cancer
View the full list of Breast Cancer Research Results and Study Updates .
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
- My Bibliography
- Collections
- Citation manager
Save citation to file
Email citation, add to collections.
- Create a new collection
- Add to an existing collection
Add to My Bibliography
Your saved search, create a file for external citation management software, your rss feed.
- Search in PubMed
- Search in NLM Catalog
- Add to Search
Connected and supported: a scoping review of how online communities provide social support for breast cancer survivors
Affiliations.
- 1 VITAM - Centre for Sustainable Health Research, Integrated University Health and Social Services Center of Capitale-Nationale, Quebec City, QC, Canada. [email protected].
- 2 Evidence-Based Practice Center, Robert D. and Patricia E. Kern Center for the Science of Health Care Delivery, Mayo Clinic, Rochester, MN, USA.
- 3 Milken Institute School of Public Health, The George Washington University, Washington, DC, USA.
- 4 Knowledge and Evaluation Research (KER) Unit, Mayo Clinic, Rochester, MN, USA.
- 5 Department of English, University of Cincinnati, Cincinnati, OH, USA.
- 6 VITAM - Centre for Sustainable Health Research, Integrated University Health and Social Services Center of Capitale-Nationale, Quebec City, QC, Canada.
- 7 Biomedical Research Institute Sant Pau (IIB Sant Pau), Barcelona, Spain.
- 8 Universidad Peruana Cayetano Heredia, Lima, Peru.
- 9 Centre Hospitalier Universitaire de Québec-Université Laval Research Centre, Quebec City, QC, Canada.
- 10 Faculty of Nursing, Laval University, Quebec City, QC, Canada.
- 11 Department of Family and Emergency Medicine, Faculty of Medicine, Université Laval, Quebec City, QC, Canada.
- PMID: 39196462
- DOI: 10.1007/s11764-024-01660-w
Purpose: To (i) assess how and to what extent online communities are used among breast cancer survivors (BCS) as a source of social support, (ii) describe the kind of support BCS access through online communities, and (iii) explore how these communities foster social support for BCS that promotes well-being and reduces the challenges of survivorship.
Methods: We conducted a scoping review. A professional librarian performed a comprehensive search in multiple databases from January 2010 to May 2023. The review process adhered to the Johana Briggs Institute's method guidelines and the PRISMA-ScR reporting system.
Results: Fifteen studies were included. Participants used social media, cancer support communities, message boards, or websites for information and emotional support. Qualitative findings resulted in four themes: to reassure; to empower; to promote equity, diversity, and inclusion; and to demonstrate for BCS the drawbacks of online support.
Conclusions: We underscore that a variety of internet websites and social media platforms are valuable for and appreciated by BCS, especially as a source of social support and human connectedness. Our study raises the existing gap in cultural/ethnic representation in this field and shows that institutional and organizational efforts are needed to address gaps in information regarding access to social support for multiethnic BCS women.
Implications for cancer survivors: This data synthesis will empower the BCS community by sharing how they can strengthen and support their peers and community via their participation in online communities that connect and support cancer survivors in healthcare spaces.
Keywords: Breast cancer; Cancer survivors; Health promotion; Online communities; Social support.
© 2024. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
PubMed Disclaimer
- Luo G, Zhang Y, Etxeberria J, Arnold M, Cai X, Hao Y, Zou H. Projections of lung cancer incidence by 2035 in 40 countries worldwide: population-based study. JMIR Public Health Surveill. 2023;9(1):e43651. - PubMed - PMC - DOI
- CCSA. Committee, Canadian Cancer Statistics. Canadian Cancer Society web page 2019.
- Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA: A Cancer J Clin. 2019;69(5):363–85.
- Dafni U, Tsourti Z, Alatsathianos I. Breast cancer statistics in the European Union: incidence and survival across European countries. Breast Care (Basel). 2019;14(6):344–53. - PubMed - DOI
- Ganz PA, Stanton AL. Psychosocial and physical health in post-treatment and extended cancer survivorship. Clinical psycho-oncology: an international perspective. West Sussex: John Wiley & Sons; 2012; p. 237–47.
Publication types
- Search in MeSH
Related information
Linkout - more resources, miscellaneous.
- NCI CPTAC Assay Portal
- Citation Manager
NCBI Literature Resources
MeSH PMC Bookshelf Disclaimer
The PubMed wordmark and PubMed logo are registered trademarks of the U.S. Department of Health and Human Services (HHS). Unauthorized use of these marks is strictly prohibited.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Eur J Breast Health
- v.18(3); 2022 Jul

Update of the 100 Most Cited Articles on Breast Cancer: A Bibliometric Analysis
Ahmet necati Şanlı.
1 Department of General Surgery, Abdulkadir Yuksel State Hospital, Gaziantep, Turkey
The aim of this study was to perform a bibliometric analysis of the 100 most cited articles related to breast cancer.
Materials and Methods:
The research was done on the Web of Science (WOS) database. Only research articles were included in the study. Results were obtained by typing the term “breast cancer” in the WOS Search box. The results were sorted according to the number of WOS core citations and all database citations, the first author of the article, the institution of the first author, publication year, article category, and countries.
The most cited article had 10236 citations. Nearly three-quarters (70%) of the articles were from the USA and most articles were published by Harvard University. Thirty-seven percent of the articles were in the medicine, general and internal medicine categories.
Conclusion:
This bibliometric analysis identified the 100 most cited research articles about breast cancer and provided a record of historical developments and trends in breast cancer research.
• The result of this research about the 100 most cited articles on breast cancer may help to understand important studies on breast cancer and shed light on future studies.
Introduction
Breast cancer is the most prevalent cancer globally, as well as a leading cause of cancer-related death among women ( 1 ). Substantial support for breast cancer awareness and research funding has helped created advances in the diagnosis and treatment of breast cancer ( 2 ). Early detection, a novel personalized approach to treatment, and a better knowledge of the disease have all contributed to an improvement in breast cancer survival rates and a steady decline in the number of deaths related with the disease ( 3 ). Current guidance on preventing and treating breast cancer, as well as what might cause it, has come mainly from information discovered from research studies ( 4 ). The most significant component of the methodological qualities of studies is associated with an increase in citations and a high impact factor of the journal in which it was published ( 5 ). To the best of our knowledge, there is only one early study that has performed a bibliometric analysis of the attributes of the 100 most cited articles about studies concerning breast cancer ( 6 ). The aim of this study was to evaluate the current status of the 100 most frequently cited articles.
Materials and Methods
A Web of Science (WOS) (Clarivate Analytics, Philadelphia, PA, United States) search was used to collect the information for this investigation. The journals indexed in the Science Citation Index Expanded (SCI-E) were included. There were no restrictions on the journals. Over 9200 of the world’s most influential publications from 178 scientific areas are now indexed in the Science Citation Index Expanded™. More than 53 million records and 1.18 billion cited references date from 1900 to the present ( 7 ).
Inclusion Criteria
The term “breast cancer” was typed into the search box of WOS basic research with the selection of all the years and the search was performed on 11.02.2022. The search produced 621,351 published articles between 1978 and 2022. As filters, English language, SCI-E scope and research article type were selected, resulting in a reduction to 376,105 articles. These were then ranked in order of citation frequency, from highest to lowest. The study was conducted by generating a shortlist of the top 100 cited publications from this search list, which were classified by journal, study category, country and location where the research was published, authors, and publication date.
Exclusion Criteria
Articles in indexes other than SCI-E, published in languages other than English, and other types of articles, such as reviews, meeting abstracts, letters, book chapters, etc., were excluded. Also, cancer statistics articles were excluded, despite receiving more citations than the included research articles.
Written informed consent was not necessary because no patient data was included in the study. The study complied with the Declaration of Helsinki.
Statistical Analysis
No inferential statistical analysis was undertaken. All the data is given in percentages, numbers and charts.
The articles included in the study are listed according to the total number of citations in the WOS database and in the all databases (WOS database, Arabic Citation Index, BIOSIS Citation Index, Chinese Science Citation Database, Data Citation Index, Russian Science Citation Index and SciELO Citation Index). According to our results, the most cited article was by Charles M. Perou and his colleagues, with 10,236 citations in the WOS database, and the least cited article was by Lisa A Carey and her colleagues, with 1,403 citations. Considering the number of publications, the most cited author was D.J. Slamon with 25,000 citations, followed by B. Fisher with 11,809 citations, T. Sorlie with 11,343 citations, Charles M. Perou with 10,236 citations, and N.K. Aaronson with 9247 citations ( Table 1 ). It was evident that all articles received more than 1000 citations and all were published between 1985 and 2021. Twelve of the most cited articles were published in 2007, and there was one publication each for 1987, 1992, 1993, 1995, 2000, 2018, and 2021 among the most cited articles ( Figure 1 ).

Distribution of the most cited articles by publication year
These most cited articles were published in 20, high-impact factor journals, with 24 articles published in the New England Journal of Medicine, 13 in Nature, 11 in the Journal of Clinical Oncology, and 11 in Science ( Table 2 ). Seventy of the studies originated from the United States of America (USA), 13 publications from the United Kingdom (UK), six from Italy and three from Canada ( Figure 2 ).

Countries from which publications originate
The articles were sourced from 51 different centers. The institution with the most publications was Harvard University with eight articles, followed by the University of Pittsburgh with six articles, the IRCCS European Institute of Oncology (IEO) with five articles, and the University of North Carolina with five articles, while 32 institutions had only one publication each ( Table 3 ).

According to WOS publication categories, 37% of the articles were in the field of medicine, general and internal medicine, medicine, research & experimental, cell biology; pathology and surgery were the least published categories in this list. In addition, when the categories we created according to the content of the articles were examined, most articles were on genetics and drug research (47% and 24%, respectively) ( Table 4 ).

Discussion and Conclusion
Citation analysis is used to find important papers on a certain subject. It aids in the analysis of scientific influence while also acknowledging substantial/pioneering contributions made by predecessors and noteworthy research advancement. There are numerous bibliometric article analyses conducted in various areas of medicine ( 8 , 9 , 10 , 11 ). To the best of our knowledge, there is only one previous article about the 100 most cited articles concerning breast cancer, and it was published in 2017 ( 6 ). Since research areas can change due to advances in science and technology, we found that the total number of citations in this study, which we aimed to evaluate the current status of the 100 most frequently cited articles, reached 280,906, an increase of approximately 1.6 times compared to 2017. This result suggests that interest in quality publications on breast cancer has increased. Also, 41 of the articles in the list were found to have changed. The vast majority of articles on the list were on chemotherapy and genetic studies.
The number of citations may be related to the time since publication. As the publication time increases, the number of citations also increases. In our study, we observed that 12 articles from 2007 and 9 articles from 2005 entered the list ( Figure 1 ). However, many factors, such as the content of the article, its quality and the journal in which it was published, can affect the number of citations. Therefore, although it was published in 2021, the study by Xi Wang and his colleagues was the fourth most cited article ( 6 ).
As expected, the most cited articles were published in the medical journals with the highest impact factors. In the present study, most articles were published in the New England Journal of Medicine, followed by articles in Nature, the Journal of Clinical Oncology, and Science, respectively. The first three articles on the list were published in Nature, Science and the Journal of the National Cancer Institute, respectively. It is feasible to hypothesize that the audience of a general medical journal is particularly interested in the topic of breast cancer, or that authors of breast cancer research choose popular medical journals to reach more researchers and readers. One of the important points in the study was that 70% of the articles originated from the USA. Similar to our study, in the bibliographic studies in the literature, 70%–93% of the research articles were USA based ( 8 , 9 , 10 , 11 ). The fact that these quality studies originate from the USA can be explained by the large patient population and the presence of many well-funded cancer centers.
The first most cited article was “Molecular portraits of human breast tumours” written by Perou et al. ( 12 ) in 2000. In this study, in which they made a molecular portrait of breast cancer, they created a molecular subtype classification of breast cancer ( 12 ). Today, this molecular classification is still in use and therefore the topic of this article remains relevant.
The second most cited article was “Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene” written by Slamon et al. ( 13 ) in 1987. In this study, they showed that the HER-2/neu oncogene may play a role in the biological behavior and pathogenesis of human breast cancer. Also, they found that amplification of the HER-2/neu gene is an important predictor of both overall survival and time to relapse in patients with breast cancer, and that the HER-2/neu oncogene plays a role in the biological behavior and pathogenesis of human breast cancer.
The third most cited article was “The European Organization for Research and Treatment of Cancer QLQ-C30: a quality-of-life instrument for use in international clinical trials in oncology” written by Aaronson et al. ( 14 ) in 1993. The EORTC QLQ-C30 questionnaire was developed as a reliable and valid measure of cancer patients’ quality of life in multicultural clinical research settings in this multicenter survey performed by the European Organization for Research and Treatment of Cancer.
The other most cited articles are in the fields of chemotherapy, gene expression, tumor genetics, pathology, and surgery. Research in these areas has made important contributions to the understanding of breast cancer. According to WOS categories, 91% of the articles were in the field of general internal medicine, multidisciplinary sciences was the second most common category, and cancer research came third. Surgery was one of the least-published fields. As the biological behavior and pathogenesis of breast cancer are better understood, studies on chemotherapy drugs have come to the fore. A possible reason for the increase in these types of studies is the increase in funding for drug research in the treatment of breast cancer. For this reason, research on surgery may have lagged behind.
Although citation analysis is a useful method that can provide insight into trends in the literature, it is not without flaws. Only the WOS database was used in this study. Thus, publications that may be indexed in other databases, such as Scopus and Google Scholar, were not included in the list of this study. Also, self-citations, lectures and textbooks were not evaluated. A search was made by typing only the term “breast cancer” in the WOS search box. Other terms that may be related to breast cancer, such as “breast, breast neoplasm, breast surgery, etc.,” were not searched. Another limitation was that the research area was examined according to the research categories determined by WOS. A more detailed investigation could not be made.
In conclusion, in this study, in which a bibliographic analysis of the 100 most cited articles in WOS on breast cancer was performed, it was observed that the number of citations increased by 1.6 times in the last 5 years. It was found that the most cited articles were published in high impact factor journals, especially the New England Journal of Medicine, most publications were from 2007, and the most cited articles were from the USA and Harvard University. Most studies focused on gene expression and chemotherapy. The result of this research may help to understand important studies on breast cancer and shed light on future studies.
Ethics Committee Approval: No ethical approval was obtained because this study did not involve a prospective evaluation, did not involve laboratory animals and only involved non-invasive procedures (e.g. faecal samples, voided urine etc).
Informed Consent: N/A
Peer-review: Externally peer-reviewed.
Financial Disclosure: The authors declared that this study received no financial support.
- Frontiers in Endocrinology
- Cancer Endocrinology
- Research Topics
Application of Nanomaterials in the Diagnosis and Treatment of Breast Cancer
Total Downloads
Total Views and Downloads
About this Research Topic
Breast cancer, the most prevalent malignancy among women globally, poses a significant threat to women's health. With over 2.3 million new cases and 670,000 deaths in 2022, advancements in treatment are urgently needed. Traditional therapies, including surgery, radiotherapy, and chemotherapy, have shown promise but face challenges due to tumor heterogeneity and drug resistance. Additionally, implant-related complications in breast reconstruction add to patient concerns. Thus, developing novel diagnostics and therapies is imperative. Nanomaterials, with their unique properties, offer potential solutions. They have demonstrated promise in breast cancer research, enhancing detection, diagnosis, and treatment. By conjugating with specific ligands/receptors, nanomaterials can improve diagnostic accuracy and enable molecular typing of breast cancer. Nanodelivery systems enhance drug targeting and stability, while nanotechnologies like phototherapy and hyperthermia provide new treatment strategies. Notably, several nanoplatforms have already been licensed for cancer therapy, with numerous others in clinical trials. However, challenges remain. Ensuring biocompatibility and safety, achieving precision targeting and smart drug release, designing multi-functional nanoplatforms, addressing production costs and clinical application, overcoming interdisciplinary research shortages, and realizing personalized treatment are urgent issues. Enhancing the performance of nanomaterials and deepening the understanding of nano-biology (tumor) are expected to drive the development of nanoscience and clinical medicine, thereby improving the current status of breast cancer diagnosis and treatment. The goal of this Research Topic is to explore the application of nanomaterials in breast cancer diagnosis and treatment, addressing key challenges such as biocompatibility, safety, precise targeting, drug loading and release efficiency, and clinical translation. By harnessing nanotechnology, we aim to develop innovative solutions that overcome these obstacles, ultimately achieving widespread and effective use of nanomaterials in breast cancer care. The current Research Topic will focus on the latest applications of nanomaterials in breast cancer diagnosis and treatment, exploring the underlying molecular mechanisms. This exploration aims to offer a fresh perspective on clinical advancements in breast cancer management and nanomaterials technology innovation. We welcome original research articles and comprehensive reviews on the following nanomaterials-related topics, among others: • Imaging Diagnosis of Breast Cancer • Detection of Breast Cancer Biomarkers • Breast Cancer Vaccines • Real-Time Navigation in Breast Cancer Surgery • Drug/Gene/Micromolecule Delivery Systems Targeting Breast Cancer • Postoperative Reconstruction for Breast Cancer.
Keywords : Nanomaterials, Breast Cancer, Endocrinology
Important Note : All contributions to this Research Topic must be within the scope of the section and journal to which they are submitted, as defined in their mission statements. Frontiers reserves the right to guide an out-of-scope manuscript to a more suitable section or journal at any stage of peer review.
Topic Editors
Topic coordinators, submission deadlines.
| Manuscript Summary | |
| Manuscript |
Participating Journals
Manuscripts can be submitted to this Research Topic via the following journals:
total views
- Demographics
No records found
total views article views downloads topic views
Top countries
Top referring sites, about frontiers research topics.
With their unique mixes of varied contributions from Original Research to Review Articles, Research Topics unify the most influential researchers, the latest key findings and historical advances in a hot research area! Find out more on how to host your own Frontiers Research Topic or contribute to one as an author.

Breast Cancer Research and Treatment
Innovative Concepts
- © 2023
- Ouissam Al Jarroudi 0 ,
- Khalid El Bairi 1 ,
- Giuseppe Curigliano 2
Department of Medical Oncology, Mohammed VI University Hospital, Oujda, Morocco
You can also search for this editor in PubMed Google Scholar
Division of New Drugs and Early Drug Development, European Institute of Oncology IRCCS, Milan, Italy
- Focuses on treatment options for breast cancer, such as radiotherapy, systemic therapy and immunotherapy
- Addresses ongoing research in screening, diagnosis and management for all subtypes of breast cancer
- Edited and authored by leading experts in the field
Part of the book series: Cancer Treatment and Research (CTAR, volume 188)
7642 Accesses
7 Citations
11 Altmetric
This is a preview of subscription content, log in via an institution to check access.
Access this book
Subscribe and save.
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
- Available as EPUB and PDF
- Read on any device
- Instant download
- Own it forever
- Durable hardcover edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
Tax calculation will be finalised at checkout
Other ways to access
Licence this eBook for your library
Institutional subscriptions
About this book
- Breast Cancer Treatment
- Triple-negative Breast Cancer
- Tumor-infiltrating Lymphocytes
- Immune Checkpoint Inhibitors
- Screening programs for breast cancer
- Precision surgery for the treatment of breast cancer
- HER2- positive breast cancer
- Biomarkers in Breast Cancer
- Luminal B breast cancer
- Onco-immunology of breast cancer
Table of contents (14 chapters)
Front matter, antibody–drug conjugates: a new therapeutic approach for triple-negative breast cancer.
- Ouissam Al Jarroudi, Khalid El Bairi, Giuseppe Curigliano, Said Afqir
Immune-Checkpoint Inhibitors: A New Line of Attack in Triple-Negative Breast Cancer
Screening programs for breast cancer: toward individualized, risk-adapted strategies of early detection.
- Dario Trapani, Josè Sandoval, Pamela Trillo Aliaga, Liliana Ascione, Pier Paolo Maria Berton Giachetti, Giuseppe Curigliano et al.
Transversal Perspectives of Integrative Oncology Care in Gastric and Lobular Breast Cancer
- Emilio Francesco Giunta, Gianluca Arrichiello, Annalisa Pappalardo, Piera Federico, Angelica Petrillo
Assessment and Response to Neoadjuvant Treatments in Breast Cancer: Current Practice, Response Monitoring, Future Approaches and Perspectives
- Vincenzo Sabatino, Alma Pignata, Marvi Valentini, Carmen Fantò, Irene Leonardi, Michela Campora
Estimating the Benefit of Preoperative Systemic Therapy to Reduce the Extent of Breast Cancer Surgery: Current Standard and Future Directions
- Giacomo Montagna
A Precise Approach for Radiotherapy of Breast Cancer
- Samantha Sigurdson, Stephane Thibodeau, Martin Korzeniowski, Fabio Ynoe Moraes
Fast Mimicking Diets and Other Innovative Nutritional Interventions to Treat Patients with Breast Cancer
- Federica Giugliano, Laura Boldrini, Jacopo Uliano, Edoardo Crimini, Ida Minchella, Giuseppe Curigliano
Mechanisms of Endocrine Resistance in Hormone Receptor-Positive Breast Cancer
- Antonio Marra, Dario Trapani, Emanuela Ferraro, Giuseppe Curigliano
Innovative Therapeutic Approaches for Patients with HER2-Positive Breast Cancer
- Beatrice Taurelli Salimbeni, Emanuela Ferraro, Luca Boscolo Bielo, Giuseppe Curigliano
Breast Cancer Brain Metastases: Achilles’ Heel in Breast Cancer Patients’ Care
- Emanuela Ferraro, Andrew D. Seidman
New Concepts in Cardio-Oncology
- Paola Zagami, Eleonora Nicolò, Chiara Corti, Carmine Valenza, Giuseppe Curigliano
Next-Generation Sequencing for Advanced Breast Cancer: What the Way to Go?
- Dario Trapani, Edoardo Crimini, José Sandoval, Giuseppe Curigliano
The Global Landscape on the Access to Cancer Medicines for Breast Cancer: The ONCOLLEGE Experience
- Csongor György Lengyel, Baker Shalal Habeeb, Sara Cecilia Altuna, Dario Trapani, Shah Zeb Khan, Sadaqat Hussain
Editors and Affiliations
Ouissam Al Jarroudi, Khalid El Bairi
Giuseppe Curigliano
About the editors
Dr. Ouissam Al Jarroudi, MD, is a distinguished medical oncologist practicing in the medical oncology department at Mohammed VI University Hospital in Oujda, Morocco. She holds a position as a professor at the Faculty of Medicine and Pharmacy, affiliated with Mohammed Ist University. Dr. Al Jarroudi has pursued various fellowships at renowned institutions such as the Department of Medical Oncology at Paul Brousse Hospital, Assistance Publique - Hopitaux de Paris, and Léon Bérard Center in France.
Dr. Al Jarroudi's research is primarily focused on prognostic and predictive biomarkers in breast cancer and she is currently a member of the European Society for Medical Oncology (ESMO) and the American Society of Clinical Oncology (ASCO).
Khalid El Bairi is a clinical research fellow and an investigator in OVANORDEST studies. He is currently pursuing clinical and translational research in medical oncology. He has published many peer-reviewedarticles in the field of predictive and prognostic cancer biomarkers to improve survival outcomes in several WoS and Medline-indexed journals. His research focuses particularly on biomarkers for digestive and gynecological cancers such as ovarian and colorectal malignancies. He is currently a member of various international scientific societies such as the European Society for Medical Oncology (ESMO), the American Society of Clinical Oncology (ASCO), the European Society of Gynaecological Oncology (ESGO), and the American Association for Cancer Research (AACR). He is also an editor and reviewer for various journals and a guest editor for several special issues on emerging topics in gynecological cancers such as platinum-resistant ovarian cancer. He is also highly interested in teaching evidence-based medicine, clinical research methods, and publishing ethics to medical and PhD students and was selected for the 70th Lindau Nobel Laureate Meeting as a young scientist. He is also involved in “global oncology” initiatives through providing free training to young researchers across LMICs. He joined the ASCO Trainee & Early Career Advisory Group as a member for the 2022-2024 term and NCODA (National Community Oncology Dispensing Association, Inc.) as an advisory member of its International Executive Council in 2023.
Giuseppe Curigliano, MD PhD, is Associate Professor of Medical Oncology at the University of Milano and the Head of the Division of Early Drug Development at the European Institute of Oncology, IRCCS, Italy. He is a clinician and researcher specializing in early drug development for patients with solid tumors with a special commitment to breast cancer. He has been a member of the Italian National Health Council since 2018 and, in 2019, he served as Chair of the Scientific Committee of The Lega Nazionale Lotta ai Tumori. He has served as a Member of the ESMO Breast Cancer Faculty since 2001 and he is currently the Faculty Coordinator. He has alsoserved on the Scientific Committee for the St Gallen Conference since 2011 and was the Scientific Co-Chair in St Gallen 2017 and 2019. He has been an Editorial Board Member for Annals of Oncology since 2014, and serves as Co-Editor in Chief of The Breast, Co-Editor in Chief of Cancer Treatment Reviews, Associate Editor of the European Journal of Cancer, Editor of the Journal of Clinical Oncology. He also serves on the European School of Oncology (ESO) faculty committee.
Bibliographic Information
Book Title : Breast Cancer Research and Treatment
Book Subtitle : Innovative Concepts
Editors : Ouissam Al Jarroudi, Khalid El Bairi, Giuseppe Curigliano
Series Title : Cancer Treatment and Research
DOI : https://doi.org/10.1007/978-3-031-33602-7
Publisher : Springer Cham
eBook Packages : Medicine , Medicine (R0)
Copyright Information : The Editor(s) (if applicable) and The Author(s), under exclusive license to Springer Nature Switzerland AG 2023
Hardcover ISBN : 978-3-031-33601-0 Published: 05 January 2024
Softcover ISBN : 978-3-031-33604-1 Due: 18 January 2025
eBook ISBN : 978-3-031-33602-7 Published: 04 January 2024
Series ISSN : 0927-3042
Series E-ISSN : 2509-8497
Edition Number : 1
Number of Pages : VIII, 368
Number of Illustrations : 5 b/w illustrations, 28 illustrations in colour
Topics : Oncology , Gynecology , Cancer Research
- Publish with us
Policies and ethics
- Find a journal
- Track your research

Elle Macpherson’s breast cancer: when the media reports on celebrity cancer, are we really getting the whole story?
NHMRC Emerging Leader Research Fellow, University of Sydney
Senior Lecturer and Coordinator, Health and Medical Humanities, University of Sydney
Professor of Clinical Epidemiology, Sydney School of Public Health, University of Sydney
Disclosure statement
Brooke Nickel receives fellowship funding from the National Health and Medical Research Council (NHMRC). She is on the Scientific Committee of the Preventing Overdiagnosis Conference.
Katy Bell receives funding from the National Health and Medical Research Council (NHMRC). She is an Investigator for Wiser Healthcare, an Australian research collaboration that aims to promote better value care for all Australians https://www.wiserhealthcare.org.au/ .
Claire Hooker does not work for, consult, own shares in or receive funding from any company or organisation that would benefit from this article, and has disclosed no relevant affiliations beyond their academic appointment.
University of Sydney provides funding as a member of The Conversation AU.
View all partners
Celebrity supermodel Elle Macpherson disclosed in an interview with The Australian Women’s Weekly earlier this week that seven years ago she was diagnosed with breast cancer.
Media coverage around the world said Macpherson had rejected some “conventional” treatments for the type of breast cancer she had disclosed, known as HER2-positive oestrogen receptive intraductal carcinoma.
This is not the first time we’ve seen powerful celebrity stories about cancer have the potential to influence the public health narrative. Sometimes these celebrity stories have changed cancer screening and treatment .
For instance, after singer Kylie Minogue announced her breast cancer diagnosis in 2005 there was an unprecedented increase in mammography bookings.
Actor Angelina Jolie’s op-ed in The New York Times in 2013 about her preventative double mastectomy for breast cancer may have inadvertently fuelled overtesting among women not at high risk.
And when actor Ben Stiller announced in 2016 that the prostate-specific antigen (PSA) test he had taken in his late 40s had saved his life, this was in contradiction to international screening guidelines . These recommend men under 55 do not use the PSA test because prostate cancer can often be overdiagnosed .
Should we be worried about the latest news?
Organisations, such as Breast Cancer Network Australia, have made public statements , worried that Macpherson’s comments might encourage an approach to treating invasive breast cancers that includes the use of non-evidence-based “wellness” products and interventions.
But media coverage of Macpherson’s situation has largely missed a key piece of information: her breast cancer is not invasive.
The type she disclosed is commonly known as ductal carcinoma in situ or DCIS . This is a cluster of pre-invasive or non-invasive breast cancer cells. It differs from invasive breast cancer in that the lesions are contained and have not spread. This means that treatments for invasive and non-invasive breast cancer differ .
In fact, Macpherson appears to have followed recommended treatment for her cancer. She did have surgery, a lumpectomy to remove the DCIS. Guidelines recommend patients weigh up the possible benefits and risks of the additional treatments that Macpherson said her doctor offered: mastectomy surgery, radiation, chemotherapy, and hormone therapy. Together with their treating team, each patient may decide whether any of these additional treatments are right for their individual situation.
There are ongoing research trials looking into who is most likely to benefit from these additional treatments, and who might not need them at all. Therefore, Macpherson’s decision to decline the additional treatments may have been both a reasonable and a conventional decision for a woman with non-invasive breast cancer.
This missing information from the media is also a missed opportunity to discuss less invasive options for the management of DCIS.
The rate of DCIS has increased greatly since the introduction of breast cancer screening. You can detect it on a mammogram but it rarely causes symptoms. Many of these lesions are unlikely to ever cause a problem in a woman’s lifetime. As a result of this some cases of DCIS are considered to be over-diagnosed .
Now options such as active surveillance (closely monitoring but not providing treatment unless the condition progresses) are considered reasonable and are being robustly evaluated in research trials to help reduce overtreatment.
We need to be wary of simplistic narratives about celebrity cancer journeys that don’t necessarily tell the whole story. This should also include scepticism over “wellness” narratives as these can lead to non-evidence-based treatment choices that waste consumers’ money and may cause them harm.
We all need to get better at being appropriately sceptical about health information without losing trust in proven health interventions.
I’m worried about my breast cancer. What should I do?
A breast cancer diagnosis can create a flood of different emotions, and presents a woman with many uncertainties including the effectiveness of treatments, and about their potential side effects and long-term impacts.
Women can ask their health professionals questions about possible management options, including:
what are my options? One of these options might be to choose less treatment, including an active surveillance approach for low-risk DCIS
what are the possible benefits and harms of those options?
how likely are each of those benefits and harms to happen to me?
The Conversation approached Elle Macpherson’s spokesperson to clarify details about her diagnosis and treatment but did not receive a response before publication.
- Public health
- Breast cancer
- Cancer treatment
- Supermodels
- Health in the news
- news analysis

Service Centre Senior Consultant

Director of STEM

Community member - Training Delivery and Development Committee (Volunteer part-time)

Chief Executive Officer

Head of Evidence to Action
- Coronavirus Updates
- Education at MUSC
- Adult Patient Care
- Children's Health
- Hollings Cancer Center
- Research at MUSC
- MUSC Catalyst News
- Triple Negative Breast Cancer
Double trouble for triple-negative breast cancer

A team of MUSC Hollings Cancer Center researchers has discovered one way in which triple-negative breast cancer (TNBC) cells become resistant to immunotherapy and have tested a two-pronged treatment strategy that was able to restore sensitivity to immunotherapy in a preclinical model. The team, led by Besim Ogretmen, Ph.D. , SmartState Endowed Chair in Lipidomics and Drug Discovery at MUSC, reports its findings in Cell Reports.
TNBC accounts for 10% to 20% of all breast cancer cases and is highly aggressive, with five-year survival rates markedly lower than for other breast cancers. It is twice as likely to occur in women under 40 than those older than 50 and is more common in Black women. TNBC is often diagnosed late, even after it has metastasized, making it difficult to treat.
TNBC earns its “triple-negative” moniker because patients with the disease already have three strikes against them. Because TNBC cells lack two necessary receptors, hormone-based therapies, a mainstay of breast cancer treatments, won’t work. Neither will therapies that specifically target the protein HER2, as patients with TNBC lack it or have only very low levels. These three strikes leave patients with TNBC, particularly metastatic TNBC, with few treatment options.
“There's not a magic bullet target for triple-negative breast cancer,” said Wyatt Wofford, an M.D., Ph.D. candidate in the Ogretmen Lab at MUSC and lead author of the Cell Reports article. “Typically, response rates for TNBC are around 15% to 20%, and so that leaves about 80% of your patients who have metastatic TNBC with no good option for therapy.”
"Use of immunotherapy by itself or the metastatic pathway inhibitor by itself did almost nothing. The cancer cells just weren’t listening. And then, when you combined these two together, there was a really pronounced response. The tumors stopped growing and even started to regress.” -- Wyatt Wofford
In recent years, immunotherapy has proved to be a breakthrough therapy for many blood cancers but has had limited success in solid tumors like TNBC. The checkpoint inhibitor pembrolizumab has been approved for recurrent TNBC for patients with at least 10% of their tumor cells expressing the immunosuppressive protein PD-L1. PD-L1 on the surface of cancer cells binds with PD-1 on the surface of immune cells known as T-cells, preventing them from do
The Ogretmen Lab studies sphingolipids, which are fat molecules that provide rigidity and stability to the cell membrane. One of the sphingolipids is ceramide. Enzymes synthesize a variety of ceramides, each with a different fatty acid chain length and a different role in cancer development and progression.
One of those enzymes, ceramide synthase 4 (CERS4), created ceramides that were important to maintaining membrane stability.
“The ceramide that this enzyme generates seems to be important to keep the tumor cell membrane intact so that everything stays where it needs to be,” said Ogretmen. “When you lose this ceramide, the PD-L1 protein doesn’t stay on the surface of the cells.”
In their study, Ogretmen and Wofford found that when levels of that enzyme dropped and not enough of the ceramides it produced were available, the membrane became unstable, enabling PD-LI to drop down inside the tumor cell.
There, hidden inside the cell, PD-L1 was not exposed to the immunotherapeutic agent as it was while on the surface. With this finding, Ogretmen and Wofford identified a mechanism by which TNBC becomes resistant to immunotherapy. They also showed that the internalized PD-L1 could promote cellular pathways linked to metastasis.
The MUSC team next wanted to know whether they could make the tumor cells once again vulnerable to immunotherapy. The researchers treated a mouse model of TNBC lacking the enzyme CERS4 with both a PD-L1 inhibitor and an existing anti-cancer drug that blocks one of the metastatic cellular pathways promoted by the internalized PD-L1.
“In this study, we were looking not only to study how TNBC becomes more resistant to immunotherapy but to use what we learned to make these cancer cells more responsive to immunotherapy." -- Dr. Besim Ogretmen
Before treatment, the mice uniformly developed lung metastases. However, after treatment, the cell membrane of the tumor regained its stability, and PD-L1 remained on the surface of the cell where it was exposed to immunotherapy and unable to promote tumor growth and metastasis.
Ogretmen and Wofford turned to Ozgur Sahin Ph.D. , SmartState Endowed Chair in the Department of Biochemistry and Molecular Biology, to do the drug studies. Wofford still remembers how excited he was when he first saw the results of these studies.
“When we looked at how the tumors were growing over time, use of immunotherapy by itself or the metastatic pathway inhibitor by itself did almost nothing. It was almost like you were giving the cancer cells water – they just weren’t listening,” he said. “And then, when you combined these two together, there was a really pronounced response. The tumors stopped growing and even started to regress.”
For Ogretmen, the discovery of a biological mechanism for resistance in TNBC, although interesting in itself, is exciting largely because it will allow researchers to manipulate that mechanism to restore sensitivity to treatment.
“In this study, we were looking not only to study how TNBC becomes more resistant to immunotherapy but to use what we learned to make these cancer cells more responsive to immunotherapy,” he said.
Currently, it is not feasible to move forward to the clinic with the pathway inhibitor tested in the study. The team’s next steps are to identify other compounds, ideally existing, approved drugs that act on the same metastatic pathway and that could be used in a combination therapy with a PD-L1 inhibitor in patients with TNBC.
“We have some new leads and exciting results,” said Ogretmen. “We are making progress at finding more viable combinations that we can actually take into the clinic.”
Wofford W, Kim J, Kim D, Janneh AH, Lee HG, Atilgan FC, Oleinik N, Kassir MF, Saatci O, Chakraborty P, Tokat UM, Gencer S, Howley B, Howe P, Mehrotra S, Sahin O, Ogretmen B. Alterations of ceramide synthesis induce PD-L1 internalization and signaling to regulate tumor metastasis and immunotherapy response. Cell Rep. 2024 Aug 27;43(8):114532. doi: 10.1016/j.celrep.2024.114532.
About the Author
Kimberly McGhee
Categories: Cancer , Research , Innovation

IMAGES
COMMENTS
Breast Cancer Research and Treatment is a comprehensive forum dedicated to all aspects of breast cancer research. The journal's focus spans across various disciplines including surgery, radiotherapy, medical oncology, endocrinology, epidemiology, immunology and cell biology. Provides an international platform for the discussion and resolution ...
Breast Cancer Res Treat 100(2):229-235). Failure to do so will result in the manuscript being returned to the author without peer review, as outlined by the editors of Breast Cancer Research and Treatment : Hayes DF, Ethier S, Lippman ME (2006) New guidelines for reporting of tumor marker studies in breast cancer research and treatment: REMARK.
Publishing options. Breast Cancer Research and Treatment is a hybrid open access journal. Once the article is accepted for publication, authors will have the option to choose how their article is published: Traditional publishing model - published articles are made available to institutions and individuals who subscribe to Breast Cancer ...
Before you submit. Now you've identified a journal to submit to, there are a few things you should be familiar with before you submit. Make sure you are submitting to the most suitable journal - Aims and scope. Understand the costs and funding options - Fees and funding. Make sure your manuscript is accurate and readable - Language editing ...
Breast Cancer Research: Home page
This guide outlines key points for preparing primary research manuscripts for submission to npj Breast Cancer. The corresponding author should be familiar with the journal's Editorial policies ...
npj Breast Cancer has a 2-year impact factor of 5.9 (2022), article downloads of 742,276 (2022) and 7 days from submission to first editorial decision (2022).
Login to your account. Email/Username. Your email address is a required field. ... Despite tremendous advances in breast cancer research and treatment over the past three decades—leading to a reduction in breast cancer mortality of over 40% in some high-income countries—gross inequities remain, with many groups being systematically left ...
Introduction. Breast cancer is the most commonly diagnosed cancer among female patients and is the leading cause of cancer-related death. 1 There were 300,590 new cases and 43,700 deaths of invasive breast cancer in the United States based on the 2023 prediction, accounting for approximately 30% of female cancers. 1 The treatments of breast cancer include surgery, chemotherapy, radiotherapy ...
Breast cancer is the most prevalent malignant tumor among women with high heterogeneity. Traditional techniques frequently struggle to comprehensively capture the intricacy and variety of cellular states and interactions within breast cancer. As global precision medicine rapidly advances, single-cell RNA sequencing (scRNA-seq) has become a highly effective technique, revolutionizing breast ...
Breast cancer commonly arises in luminal cells of the mammary gland expressing the estrogen receptor (ER). The expression of ER in the tumor is a cornerstone for the selection of adjuvant treatment.
NCI is funding a large-scale randomized breast screening trial, the Tomosynthesis Mammographic Imaging Screening Trial (TMIST), to compare the number of advanced cancers detected in women screened for 5 years with 3-D mammography with the number detected in women screened with 2-D mammography. Two concerns in breast cancer screening, as in all ...
1. Introduction. Breast cancer has a very long history as it was first reported by the ancient Egyptians more than 3500 years ago in about 1500 B.C [].Today, breast cancer is the second most prevalent type of cancer and is a leading cause of most cancer-related deaths in women in the United States [].Around 281,550 women are projected to be diagnosed with breast cancer in 2021, and 43,600 ...
Breast Cancer: Basic and Clinical Research is an international, peer-reviewed, open access journal that covers all aspects of research and treatment of breast cancer. The journal aims to promote understanding of breast cancer biology and … | View full journal description. This journal is a member of the Committee on Publication Ethics (COPE).
A large study among women in sub-Saharan Africa showed that early cancer diagnosis and a multimodal treatment approach (involving surgery and systemic therapy) could prevent 28%-37% of breast cancer deaths in this underserved region. 8 Furthermore, consistent adherence to breast cancer treatment protocols can narrow or even eliminate the gap ...
The estimated new breast cancer cases reached 2.3 million in 2020, accounting for 11.7% of all new cancers, and 684,996 cases died of it . In China, breast cancer was the most common malignancy among women, with an estimated number of 306,000 new cases occurring in 2016 . The incidence of breast cancer has increased since the widespread uptake ...
Patient characteristics and treatment patterns of patients with locally advanced or metastatic HER2-low breast cancer, a single site descriptive study. Connor Willis. Chia Jie Tan. David Stenehjem. Research Open access 22 August 2024.
Breast Cancer: Targets and Therapy is an international, peer reviewed, open access journal focusing on breast cancer, basic and translational research, clinical trials and treatment. The latter includes outcome studies that incorporate preventative and integrated treatment strategies for enhanced survival, and quality of life for cancer ...
1 Introduction. Metastasis is one of the leading reasons for cancer recurrence, treatment failure, and poor prognosis, especially in triple-negative breast cancer patients. [] Triple-negative breast cancer cells are characterized with multiple resistant mechanisms, high invasive nature, and immune evasion properties. [] Therefore, commonly used surgery and radiation therapy are difficult to ...
Purpose: To (i) assess how and to what extent online communities are used among breast cancer survivors (BCS) as a source of social support, (ii) describe the kind of support BCS access through online communities, and (iii) explore how these communities foster social support for BCS that promotes well-being and reduces the challenges of survivorship.
Introduction. Hotspots (HS) mutations in the PIK3CA gene may lead to poorer oncological outcomes and endocrine resistance in advanced breast cancer (BC), but their prognostic role in early-stage disease remains controversial. The overall agreement within plasma and tissue methods has not been well explored. Our aim was to correlate tissue and plasma approaches and to analyze the prognostic ...
The most recent global cancer burden figures estimate that there were 2.26 million incident breast cancer cases in 2020 and the disease is the leading cause of cancer mortality in women worldwide.
Introduction. Breast cancer is the most prevalent cancer globally, as well as a leading cause of cancer-related death among women ().). Substantial support for breast cancer awareness and research funding has helped created advances in the diagnosis and treatment of breast cancer ().Early detection, a novel personalized approach to treatment, and a better knowledge of the disease have all ...
Introduction. Therapy with curative intent for breast cancer achieves optimal outcomes when it is administered to completion within a defined timeframe; its success depends on appropriate referrals for timely and personalized multimodality treatment after the receipt of a definitive diagnosis. 1 The provision of cancer treatment requires an organized, multidisciplinary approach in which ...
Breast cancer, the most prevalent malignancy among women globally, poses a significant threat to women's health. With over 2.3 million new cases and 670,000 deaths in 2022, advancements in treatment are urgently needed. Traditional therapies, including surgery, radiotherapy, and chemotherapy, have shown promise but face challenges due to tumor heterogeneity and drug resistance.
Focuses on treatment options for breast cancer, such as radiotherapy, systemic therapy and immunotherapy. Addresses ongoing research in screening, diagnosis and management for all subtypes of breast cancer. Edited and authored by leading experts in the field. Part of the book series: Cancer Treatment and Research (CTAR, volume 188) 7533 Accesses.
Impressive gains in breast cancer research and treatment have been made over the past 45 years in high-income countries. Many women are now cured with a simple lumpectomy, minimal lymph node surgery, and targeted or endocrine therapy. 1 However, this progress is in stark contrast to what exists in low- and middle-income countries (LMICs) 2 where women have a higher burden of breast cancer ...
Other aptamers for cancer treatment have been described but are currently at early stages of development. For example, it has recently been reported an aptamer targeting EPH receptor A2 (EphA2) [Citation 14]. EphA2 is overexpressed in many solid tumors and several modalities have been developed to target it (e.g. antibodies, immunotherapy, RNAi ...
Organisations, such as Breast Cancer Network Australia, have made public statements, worried that Macpherson's comments might encourage an approach to treating invasive breast cancers that ...
These three strikes leave patients with TNBC, particularly metastatic TNBC, with few treatment options. "There's not a magic bullet target for triple-negative breast cancer," said Wyatt Wofford, an M.D., Ph.D. candidate in the Ogretmen Lab at MUSC and lead author of the Cell Reports article.