Optimization Methods ¶
Until now, you've always used Gradient Descent to update the parameters and minimize the cost. In this notebook, you will learn more advanced optimization methods that can speed up learning and perhaps even get you to a better final value for the cost function. Having a good optimization algorithm can be the difference between waiting days vs. just a few hours to get a good result.

Notations : As usual, $\frac{\partial J}{\partial a } = $ da for any variable a .
To get started, run the following code to import the libraries you will need.

Updates to Assignment
If you were working on a previous version ¶.
- The current notebook filename is version "Optimization_methods_v1b".
- You can find your work in the file directory as version "Optimization methods'.
- To see the file directory, click on the Coursera logo at the top left of the notebook.
List of Updates ¶
- op_utils is now opt_utils_v1a. Assertion statement in initialize_parameters is fixed.
- opt_utils_v1a: compute_cost function now accumulates total cost of the batch without taking the average (average is taken for entire epoch instead).
- In model function, the total cost per mini-batch is accumulated, and the average of the entire epoch is taken as the average cost. So the plot of the cost function over time is now a smooth downward curve instead of an oscillating curve.
- Print statements used to check each function are reformatted, and 'expected output` is reformatted to match the format of the print statements (for easier visual comparisons).
1 - Gradient Descent ¶
A simple optimization method in machine learning is gradient descent (GD). When you take gradient steps with respect to all $m$ examples on each step, it is also called Batch Gradient Descent.
Warm-up exercise : Implement the gradient descent update rule. The gradient descent rule is, for $l = 1, ..., L$: $$ W^{[l]} = W^{[l]} - \alpha \text{ } dW^{[l]} \tag{1}$$ $$ b^{[l]} = b^{[l]} - \alpha \text{ } db^{[l]} \tag{2}$$
where L is the number of layers and $\alpha$ is the learning rate. All parameters should be stored in the parameters dictionary. Note that the iterator l starts at 0 in the for loop while the first parameters are $W^{[1]}$ and $b^{[1]}$. You need to shift l to l+1 when coding.
Expected Output :
A variant of this is Stochastic Gradient Descent (SGD), which is equivalent to mini-batch gradient descent where each mini-batch has just 1 example. The update rule that you have just implemented does not change. What changes is that you would be computing gradients on just one training example at a time, rather than on the whole training set. The code examples below illustrate the difference between stochastic gradient descent and (batch) gradient descent.
- (Batch) Gradient Descent :
- Stochastic Gradient Descent :
In Stochastic Gradient Descent, you use only 1 training example before updating the gradients. When the training set is large, SGD can be faster. But the parameters will "oscillate" toward the minimum rather than converge smoothly. Here is an illustration of this:

Note also that implementing SGD requires 3 for-loops in total:
- Over the number of iterations
- Over the $m$ training examples
- Over the layers (to update all parameters, from $(W^{[1]},b^{[1]})$ to $(W^{[L]},b^{[L]})$)
In practice, you'll often get faster results if you do not use neither the whole training set, nor only one training example, to perform each update. Mini-batch gradient descent uses an intermediate number of examples for each step. With mini-batch gradient descent, you loop over the mini-batches instead of looping over individual training examples.

What you should remember :
- The difference between gradient descent, mini-batch gradient descent and stochastic gradient descent is the number of examples you use to perform one update step.
- You have to tune a learning rate hyperparameter $\alpha$.
- With a well-turned mini-batch size, usually it outperforms either gradient descent or stochastic gradient descent (particularly when the training set is large).
2 - Mini-Batch Gradient descent ¶
Let's learn how to build mini-batches from the training set (X, Y).
There are two steps:
- Shuffle : Create a shuffled version of the training set (X, Y) as shown below. Each column of X and Y represents a training example. Note that the random shuffling is done synchronously between X and Y. Such that after the shuffling the $i^{th}$ column of X is the example corresponding to the $i^{th}$ label in Y. The shuffling step ensures that examples will be split randomly into different mini-batches.

- Partition : Partition the shuffled (X, Y) into mini-batches of size mini_batch_size (here 64). Note that the number of training examples is not always divisible by mini_batch_size . The last mini batch might be smaller, but you don't need to worry about this. When the final mini-batch is smaller than the full mini_batch_size , it will look like this:

Exercise : Implement random_mini_batches . We coded the shuffling part for you. To help you with the partitioning step, we give you the following code that selects the indexes for the $1^{st}$ and $2^{nd}$ mini-batches:
Note that the last mini-batch might end up smaller than mini_batch_size=64 . Let $\lfloor s \rfloor$ represents $s$ rounded down to the nearest integer (this is math.floor(s) in Python). If the total number of examples is not a multiple of mini_batch_size=64 then there will be $\lfloor \frac{m}{mini\_batch\_size}\rfloor$ mini-batches with a full 64 examples, and the number of examples in the final mini-batch will be ($m-mini_\_batch_\_size \times \lfloor \frac{m}{mini\_batch\_size}\rfloor$).
| **shape of the 1st mini_batch_X** | (12288, 64) |
| **shape of the 2nd mini_batch_X** | (12288, 64) |
| **shape of the 3rd mini_batch_X** | (12288, 20) |
| **shape of the 1st mini_batch_Y** | (1, 64) |
| **shape of the 2nd mini_batch_Y** | (1, 64) |
| **shape of the 3rd mini_batch_Y** | (1, 20) |
| **mini batch sanity check** | [ 0.90085595 -0.7612069 0.2344157 ] |
- Shuffling and Partitioning are the two steps required to build mini-batches
- Powers of two are often chosen to be the mini-batch size, e.g., 16, 32, 64, 128.
3 - Momentum ¶
Because mini-batch gradient descent makes a parameter update after seeing just a subset of examples, the direction of the update has some variance, and so the path taken by mini-batch gradient descent will "oscillate" toward convergence. Using momentum can reduce these oscillations.
Momentum takes into account the past gradients to smooth out the update. We will store the 'direction' of the previous gradients in the variable $v$. Formally, this will be the exponentially weighted average of the gradient on previous steps. You can also think of $v$ as the "velocity" of a ball rolling downhill, building up speed (and momentum) according to the direction of the gradient/slope of the hill.

**Figure 3** : The red arrows shows the direction taken by one step of mini-batch gradient descent with momentum. The blue points show the direction of the gradient (with respect to the current mini-batch) on each step. Rather than just following the gradient, we let the gradient influence $v$ and then take a step in the direction of $v$.
Exercise : Initialize the velocity. The velocity, $v$, is a python dictionary that needs to be initialized with arrays of zeros. Its keys are the same as those in the grads dictionary, that is: for $l =1,...,L$:
Note that the iterator l starts at 0 in the for loop while the first parameters are v["dW1"] and v["db1"] (that's a "one" on the superscript). This is why we are shifting l to l+1 in the for loop.
Exercise : Now, implement the parameters update with momentum. The momentum update rule is, for $l = 1, ..., L$:
where L is the number of layers, $\beta$ is the momentum and $\alpha$ is the learning rate. All parameters should be stored in the parameters dictionary. Note that the iterator l starts at 0 in the for loop while the first parameters are $W^{[1]}$ and $b^{[1]}$ (that's a "one" on the superscript). So you will need to shift l to l+1 when coding.
- The velocity is initialized with zeros. So the algorithm will take a few iterations to "build up" velocity and start to take bigger steps.
- If $\beta = 0$, then this just becomes standard gradient descent without momentum.
How do you choose $\beta$?
- The larger the momentum $\beta$ is, the smoother the update because the more we take the past gradients into account. But if $\beta$ is too big, it could also smooth out the updates too much.
- Common values for $\beta$ range from 0.8 to 0.999. If you don't feel inclined to tune this, $\beta = 0.9$ is often a reasonable default.
- Tuning the optimal $\beta$ for your model might need trying several values to see what works best in term of reducing the value of the cost function $J$.
- Momentum takes past gradients into account to smooth out the steps of gradient descent. It can be applied with batch gradient descent, mini-batch gradient descent or stochastic gradient descent.
- You have to tune a momentum hyperparameter $\beta$ and a learning rate $\alpha$.
4 - Adam ¶
Adam is one of the most effective optimization algorithms for training neural networks. It combines ideas from RMSProp (described in lecture) and Momentum.
How does Adam work?
- It calculates an exponentially weighted average of past gradients, and stores it in variables $v$ (before bias correction) and $v^{corrected}$ (with bias correction).
- It calculates an exponentially weighted average of the squares of the past gradients, and stores it in variables $s$ (before bias correction) and $s^{corrected}$ (with bias correction).
- It updates parameters in a direction based on combining information from "1" and "2".
The update rule is, for $l = 1, ..., L$:
- t counts the number of steps taken of Adam
- L is the number of layers
- $\beta_1$ and $\beta_2$ are hyperparameters that control the two exponentially weighted averages.
- $\alpha$ is the learning rate
- $\varepsilon$ is a very small number to avoid dividing by zero
As usual, we will store all parameters in the parameters dictionary
Exercise : Initialize the Adam variables $v, s$ which keep track of the past information.
Instruction : The variables $v, s$ are python dictionaries that need to be initialized with arrays of zeros. Their keys are the same as for grads , that is: for $l = 1, ..., L$:
Exercise : Now, implement the parameters update with Adam. Recall the general update rule is, for $l = 1, ..., L$:
Note that the iterator l starts at 0 in the for loop while the first parameters are $W^{[1]}$ and $b^{[1]}$. You need to shift l to l+1 when coding.
You now have three working optimization algorithms (mini-batch gradient descent, Momentum, Adam). Let's implement a model with each of these optimizers and observe the difference.
5 - Model with different optimization algorithms ¶
Lets use the following "moons" dataset to test the different optimization methods. (The dataset is named "moons" because the data from each of the two classes looks a bit like a crescent-shaped moon.)
We have already implemented a 3-layer neural network. You will train it with:
- update_parameters_with_gd()
- initialize_velocity() and update_parameters_with_momentum()
- initialize_adam() and update_parameters_with_adam()
You will now run this 3 layer neural network with each of the 3 optimization methods.
5.1 - Mini-batch Gradient descent ¶
Run the following code to see how the model does with mini-batch gradient descent.
5.2 - Mini-batch gradient descent with momentum ¶
Run the following code to see how the model does with momentum. Because this example is relatively simple, the gains from using momemtum are small; but for more complex problems you might see bigger gains.
5.3 - Mini-batch with Adam mode ¶
Run the following code to see how the model does with Adam.
5.4 - Summary ¶
| **optimization method** | **accuracy** | **cost shape** | Gradient descent | 79.7% | oscillations |
| Momentum | 79.7% | oscillations |
| Adam | 94% | smoother |
Momentum usually helps, but given the small learning rate and the simplistic dataset, its impact is almost negligeable. Also, the huge oscillations you see in the cost come from the fact that some minibatches are more difficult thans others for the optimization algorithm.
Adam on the other hand, clearly outperforms mini-batch gradient descent and Momentum. If you run the model for more epochs on this simple dataset, all three methods will lead to very good results. However, you've seen that Adam converges a lot faster.
Some advantages of Adam include:
- Relatively low memory requirements (though higher than gradient descent and gradient descent with momentum)
- Usually works well even with little tuning of hyperparameters (except $\alpha$)
References :
- Adam paper: https://arxiv.org/pdf/1412.6980.pdf

Optimization for Decision Science
Optimization for decision science #.
Computational optimization is essential to many subdomains of engineering, science, and business such as design, control, operations, supply chains, game theory, data science/analytics, and machine learning.
This course provides a practical introduction to models, algorithms, and modern software for large-scale numerical optimization , especially for decision-making in engineering and business contexts. Topics include (nonconvex) nonlinear programming, deterministic global optimization, integer programming, dynamic optimization, and stochastic programming. Multi-objective optimization, optimization with embedded machine learning models as constraints, optimal experiment design, optimization for statistical inference, and mathematical programs with complementarity constraints may be covered based on time and student interests. The class is designed for advanced undergraduate/graduate engineering, science, mathematics, business, and statistics students who wish to incorporate computation optimization methods into their research. The course begins with an introduction to modeling and the Python-based Pyomo computational environment. Optimization theory and algorithms are emphasized throughout the semester.
What am I going to get out of this class? #
At the end of the semester, you should be able to…
Mathematically formulate optimization problems relevant to decision-making within your discipline (and research!)
Program optimization models in Pyomo and compute numerical solutions with state-of-the-art (e.g., commercial) solvers
Explain the main theory for nonlinear constrained and unconstrained optimization
Describe basic algorithmic elements in pseudocode and implement them in Python
Analyze results from an optimization problem and communicate key findings in a presentation
Write and debug 200 lines of Python code using best practices (e.g., publication quality figures, doc strings)
What do I need to know to take the class? #
Graduate students (60499) : A background in linear algebra and numerical methods is strongly recommended but not required. Students must be comfortable programming in Python (preferred), MATLAB, Julia, C, or a similar language. Topics in EE 60551 and ACMS 60880 are complementary to CBE/ACMS 60499. These courses are not prerequisites for CBE/ACMS 60499.
Undergraduate students (40499) : Students must be comfortable with linear algebra and programming in Python (preferred), MATLAB, Julia, C, or a similar language. A course on these topics, such as CBE 20258, AME 30251 (concurrent is okay), CE 30125 (concurrent is okay), PHYS 20451, or ACMS 20220, and the standard undergraduate curriculum in computer science or electrical engineering should satisfy this prerequisite. Please contact the instructor for any preparation questions.
Organization
- Assignments
Optimization Modeling in Pyomo
- 1.1. Local Installation
- 1.2. Optimization Modeling with Applications
- 1.3. Your First Optimization Problem
- 1.4. Continuous Optimization
- 1.5. Integer Programs
- 1.6. 60 Minutes to Pyomo: An Energy Storage Model Predictive Control Example
- 2.1. Logical Modeling and Generalized Disjunctive Programs
- 2.2. Modeling Disjunctions through the Strip Packing Problem
- 3.1. Pyomo.DAE Example: Race Car
- 3.2. Pyomo.DAE Example: Temperature Control Lab
- 3.3. Differential Algebraic Equations (DAEs)
- 3.4. Numeric Integration for DAEs
- 3.5. Dynamic Optimization with Collocation and Pyomo.DAE
- 3.6. Pyomo.DAE: Racing Example Revisited
- 4.1. Stochastic Programming
- 4.2. Risk Measures and Portfolio Optimization
- 5.1. Parameter estimation with parmest
- 5.2. Supplementary material: data for parmest tutorial
- 5.3. Reactor Kinetics Example for Pyomo.DoE Tutorial
Algorithms and Theory
- 6.1. Linear Algebra Review and SciPy Basics
- 6.2. Mathematics Primer
- 6.3. Unconstrained Optimality Conditions
- 6.4. Newton-type Methods for Unconstrained Optimization
- 6.5. Quasi-Newton Methods for Unconstrained Optimization
- 6.6. Descent and Globalization
- 7.1. Convexity Revisited
- 7.2. Local Optimality Conditions
- 7.3. Analysis of KKT Conditions
- 7.4. Constraint Qualifications
- 7.5. Second Order Optimality Conditions
- 7.6. NLP Diagnostics with Degeneracy Hunter
- 7.7. Simple Netwon Method for Equality Constrained NLPs
- 7.8. Inertia-Corrected Netwon Method for Equality Constrained NLPs
- 8.1. Integer Programming with Simple Branch and Bound
- 8.2. MINLP Algorithms
- 8.3. Deterministic Global Optimization
Student Contributions
- Bayesian Optimization Tutorial 1
- Bayesian Optimization Tutorial 2
- Stochastic Gradient Descent Tutorial 1
- Stochastic Gradient Descent Tutorial 2
- Functions and Utilities
- Implementation of the Algorithm
- Regression Example: Fitting a Quadratic Function to a 2D Data Set
- Binary Classification: Logistic Regression Example
- Conclusions
- Stochastic Gradient Descent Tutorial 3
- Expectation Maximization Algorithm and MAP Estimation

Optimization in Machine Learning
This website offers an open and free introductory course on optimization for machine learning. The course is constructed holistically and as self-contained as possible, in order to cover most optimization principles and methods that are relevant for optimization.
This course is recommended as an introductory graduate-level course for Master’s level students.
If you want to learn more about this course, please (1) read the outline further below and (2) read the section on prerequisites
Later on, please note: (1) The course uses a unified mathematical notation. We provide cheat sheets to summarize the most important symbols and concepts. (2) Most sections already contain exercises with worked-out solutions to enable self-study as much as possible.
The course material is developed in a public github repository: https://github.com/slds-lmu/lecture_optimization .
If you love teaching ML and have free resources available, please consider joining the team and email us now! ( [email protected] or [email protected] )
- Chapter 1.1: Differentiability
- Chapter 1.2: Taylor Approximation
- Chapter 1.3: Convexity
- Chapter 1.4: Conditions for optimality
- Chapter 1.5: Quadratic forms I
- Chapter 1.6: Quadratic forms II
- Chapter 1.7: Matrix calculus
- Chapter 2.1: Unconstrained optimization problems
- Chapter 2.2: Constrained optimization problems
- Chapter 2.3: Other optimization problems
- Chapter 3.1: Golden ratio
- Chapter 3.2: Brent
- Chapter 4.01: Gradient descent
- Chapter 4.02: Step size and optimality
- Chapter 4.03: Deep dive: Gradient descent
- Chapter 4.04: Weaknesses of GD – Curvature
- Chapter 4.05: GD – Multimodality and Saddle points
- Chapter 4.06: GD with Momentum
- Chapter 4.07: GD in quadratic forms
- Chapter 4.09: SGD
- Chapter 4.10: SGD Further Details
- Chapter 4.11: ADAM and friends
- Chapter 5.01: Newton-Raphson
- Chapter 5.03: Gauss-Newton
- Chapter 6.01: Introduction
- Chapter 6.02: Linear Programming
- Chapter 6.04: Duality in optimization
- Chapter 6.05: Nonlinear programs and Lagrangian
- Chapter 7.01: Coordinate Descent
- Chapter 7.02: Nelder-Mead
- Chapter 7.03: Simulated Annealing
- Chapter 7.04: Multi-Starts
- Chapter 8.01: Introduction
- Chapter 8.02: ES / Numerical Encodings
- Chapter 8.03: GA / Bit Strings
- Chapter 8.04: CMA-ES Algorithm
- Chapter 8.05: CMA-ES Algorithm Wrap Up
- Chapter 10.01: Black Box Optimization
- Chapter 10.02: Basic BO Loop and Surrogate Modelling
- Chapter 10.03: Posterior Uncertainty and Acquisition Functions I
- Chapter 10.04: Posterior Uncertainty and Acquisition Functions II
- Chapter 10.05: Important Surrogate Models
- Cheat Sheets
Optimization ( scipy.optimize ) #
The scipy.optimize package provides several commonly used optimization algorithms. A detailed listing is available: scipy.optimize (can also be found by help(scipy.optimize) ).
Local minimization of multivariate scalar functions ( minimize ) #
The minimize function provides a common interface to unconstrained and constrained minimization algorithms for multivariate scalar functions in scipy.optimize . To demonstrate the minimization function, consider the problem of minimizing the Rosenbrock function of \(N\) variables:
The minimum value of this function is 0 which is achieved when \(x_{i}=1.\)
Note that the Rosenbrock function and its derivatives are included in scipy.optimize . The implementations shown in the following sections provide examples of how to define an objective function as well as its jacobian and hessian functions. Objective functions in scipy.optimize expect a numpy array as their first parameter which is to be optimized and must return a float value. The exact calling signature must be f(x, *args) where x represents a numpy array and args a tuple of additional arguments supplied to the objective function.
Unconstrained minimization #
Nelder-mead simplex algorithm ( method='nelder-mead' ) #.
In the example below, the minimize routine is used with the Nelder-Mead simplex algorithm (selected through the method parameter):
The simplex algorithm is probably the simplest way to minimize a fairly well-behaved function. It requires only function evaluations and is a good choice for simple minimization problems. However, because it does not use any gradient evaluations, it may take longer to find the minimum.
Another optimization algorithm that needs only function calls to find the minimum is Powell ’s method available by setting method='powell' in minimize .
To demonstrate how to supply additional arguments to an objective function, let us minimize the Rosenbrock function with an additional scaling factor a and an offset b :
Again using the minimize routine this can be solved by the following code block for the example parameters a=0.5 and b=1 .
As an alternative to using the args parameter of minimize , simply wrap the objective function in a new function that accepts only x . This approach is also useful when it is necessary to pass additional parameters to the objective function as keyword arguments.
Another alternative is to use functools.partial .
Broyden-Fletcher-Goldfarb-Shanno algorithm ( method='BFGS' ) #
In order to converge more quickly to the solution, this routine uses the gradient of the objective function. If the gradient is not given by the user, then it is estimated using first-differences. The Broyden-Fletcher-Goldfarb-Shanno (BFGS) method typically requires fewer function calls than the simplex algorithm even when the gradient must be estimated.
To demonstrate this algorithm, the Rosenbrock function is again used. The gradient of the Rosenbrock function is the vector:
This expression is valid for the interior derivatives. Special cases are
A Python function which computes this gradient is constructed by the code-segment:
This gradient information is specified in the minimize function through the jac parameter as illustrated below.
Avoiding Redundant Calculation
It is common for the objective function and its gradient to share parts of the calculation. For instance, consider the following problem.
Here, expensive is called 12 times: six times in the objective function and six times from the gradient. One way of reducing redundant calculations is to create a single function that returns both the objective function and the gradient.
When we call minimize, we specify jac==True to indicate that the provided function returns both the objective function and its gradient. While convenient, not all scipy.optimize functions support this feature, and moreover, it is only for sharing calculations between the function and its gradient, whereas in some problems we will want to share calculations with the Hessian (second derivative of the objective function) and constraints. A more general approach is to memoize the expensive parts of the calculation. In simple situations, this can be accomplished with the functools.lru_cache wrapper.
Newton-Conjugate-Gradient algorithm ( method='Newton-CG' ) #
Newton-Conjugate Gradient algorithm is a modified Newton’s method and uses a conjugate gradient algorithm to (approximately) invert the local Hessian [NW] . Newton’s method is based on fitting the function locally to a quadratic form:
where \(\mathbf{H}\left(\mathbf{x}_{0}\right)\) is a matrix of second-derivatives (the Hessian). If the Hessian is positive definite then the local minimum of this function can be found by setting the gradient of the quadratic form to zero, resulting in
The inverse of the Hessian is evaluated using the conjugate-gradient method. An example of employing this method to minimizing the Rosenbrock function is given below. To take full advantage of the Newton-CG method, a function which computes the Hessian must be provided. The Hessian matrix itself does not need to be constructed, only a vector which is the product of the Hessian with an arbitrary vector needs to be available to the minimization routine. As a result, the user can provide either a function to compute the Hessian matrix, or a function to compute the product of the Hessian with an arbitrary vector.
Full Hessian example
The Hessian of the Rosenbrock function is
if \(i,j\in\left[1,N-2\right]\) with \(i,j\in\left[0,N-1\right]\) defining the \(N\times N\) matrix. Other non-zero entries of the matrix are
For example, the Hessian when \(N=5\) is
The code which computes this Hessian along with the code to minimize the function using Newton-CG method is shown in the following example:
Hessian product example
For larger minimization problems, storing the entire Hessian matrix can consume considerable time and memory. The Newton-CG algorithm only needs the product of the Hessian times an arbitrary vector. As a result, the user can supply code to compute this product rather than the full Hessian by giving a hess function which take the minimization vector as the first argument and the arbitrary vector as the second argument (along with extra arguments passed to the function to be minimized). If possible, using Newton-CG with the Hessian product option is probably the fastest way to minimize the function.
In this case, the product of the Rosenbrock Hessian with an arbitrary vector is not difficult to compute. If \(\mathbf{p}\) is the arbitrary vector, then \(\mathbf{H}\left(\mathbf{x}\right)\mathbf{p}\) has elements:
Code which makes use of this Hessian product to minimize the Rosenbrock function using minimize follows:
According to [NW] p. 170 the Newton-CG algorithm can be inefficient when the Hessian is ill-conditioned because of the poor quality search directions provided by the method in those situations. The method trust-ncg , according to the authors, deals more effectively with this problematic situation and will be described next.
Trust-Region Newton-Conjugate-Gradient Algorithm ( method='trust-ncg' ) #
The Newton-CG method is a line search method: it finds a direction of search minimizing a quadratic approximation of the function and then uses a line search algorithm to find the (nearly) optimal step size in that direction. An alternative approach is to, first, fix the step size limit \(\Delta\) and then find the optimal step \(\mathbf{p}\) inside the given trust-radius by solving the following quadratic subproblem:
The solution is then updated \(\mathbf{x}_{k+1} = \mathbf{x}_{k} + \mathbf{p}\) and the trust-radius \(\Delta\) is adjusted according to the degree of agreement of the quadratic model with the real function. This family of methods is known as trust-region methods. The trust-ncg algorithm is a trust-region method that uses a conjugate gradient algorithm to solve the trust-region subproblem [NW] .
Trust-Region Truncated Generalized Lanczos / Conjugate Gradient Algorithm ( method='trust-krylov' ) #
Similar to the trust-ncg method, the trust-krylov method is a method suitable for large-scale problems as it uses the hessian only as linear operator by means of matrix-vector products. It solves the quadratic subproblem more accurately than the trust-ncg method.
This method wraps the [TRLIB] implementation of the [GLTR] method solving exactly a trust-region subproblem restricted to a truncated Krylov subspace. For indefinite problems it is usually better to use this method as it reduces the number of nonlinear iterations at the expense of few more matrix-vector products per subproblem solve in comparison to the trust-ncg method.
F. Lenders, C. Kirches, A. Potschka: “trlib: A vector-free implementation of the GLTR method for iterative solution of the trust region problem”, arXiv:1611.04718
N. Gould, S. Lucidi, M. Roma, P. Toint: “Solving the Trust-Region Subproblem using the Lanczos Method”, SIAM J. Optim., 9(2), 504–525, (1999). DOI:10.1137/S1052623497322735
Trust-Region Nearly Exact Algorithm ( method='trust-exact' ) #
All methods Newton-CG , trust-ncg and trust-krylov are suitable for dealing with large-scale problems (problems with thousands of variables). That is because the conjugate gradient algorithm approximately solve the trust-region subproblem (or invert the Hessian) by iterations without the explicit Hessian factorization. Since only the product of the Hessian with an arbitrary vector is needed, the algorithm is specially suited for dealing with sparse Hessians, allowing low storage requirements and significant time savings for those sparse problems.
For medium-size problems, for which the storage and factorization cost of the Hessian are not critical, it is possible to obtain a solution within fewer iteration by solving the trust-region subproblems almost exactly. To achieve that, a certain nonlinear equations is solved iteratively for each quadratic subproblem [CGT] . This solution requires usually 3 or 4 Cholesky factorizations of the Hessian matrix. As the result, the method converges in fewer number of iterations and takes fewer evaluations of the objective function than the other implemented trust-region methods. The Hessian product option is not supported by this algorithm. An example using the Rosenbrock function follows:
J. Nocedal, S.J. Wright “Numerical optimization.” 2nd edition. Springer Science (2006).
Conn, A. R., Gould, N. I., & Toint, P. L. “Trust region methods”. Siam. (2000). pp. 169-200.
Constrained minimization #
The minimize function provides several algorithms for constrained minimization, namely 'trust-constr' , 'SLSQP' , 'COBYLA' , and 'COBYQA' . They require the constraints to be defined using slightly different structures. The methods 'trust-constr' and 'COBYQA' require the constraints to be defined as a sequence of objects LinearConstraint and NonlinearConstraint . Methods 'SLSQP' and 'COBYLA' , on the other hand, require constraints to be defined as a sequence of dictionaries, with keys type , fun and jac .
As an example let us consider the constrained minimization of the Rosenbrock function:
This optimization problem has the unique solution \([x_0, x_1] = [0.4149,~ 0.1701]\) , for which only the first and fourth constraints are active.
Trust-Region Constrained Algorithm ( method='trust-constr' ) #
The trust-region constrained method deals with constrained minimization problems of the form:
When \(c^l_j = c^u_j\) the method reads the \(j\) -th constraint as an equality constraint and deals with it accordingly. Besides that, one-sided constraint can be specified by setting the upper or lower bound to np.inf with the appropriate sign.
The implementation is based on [EQSQP] for equality-constraint problems and on [TRIP] for problems with inequality constraints. Both are trust-region type algorithms suitable for large-scale problems.
Defining Bounds Constraints
The bound constraints \(0 \leq x_0 \leq 1\) and \(-0.5 \leq x_1 \leq 2.0\) are defined using a Bounds object.
Defining Linear Constraints
The constraints \(x_0 + 2 x_1 \leq 1\) and \(2 x_0 + x_1 = 1\) can be written in the linear constraint standard format:
and defined using a LinearConstraint object.
Defining Nonlinear Constraints The nonlinear constraint:
with Jacobian matrix:
and linear combination of the Hessians:
is defined using a NonlinearConstraint object.
Alternatively, it is also possible to define the Hessian \(H(x, v)\) as a sparse matrix,
or as a LinearOperator object.
When the evaluation of the Hessian \(H(x, v)\) is difficult to implement or computationally infeasible, one may use HessianUpdateStrategy . Currently available strategies are BFGS and SR1 .
Alternatively, the Hessian may be approximated using finite differences.
The Jacobian of the constraints can be approximated by finite differences as well. In this case, however, the Hessian cannot be computed with finite differences and needs to be provided by the user or defined using HessianUpdateStrategy .
Solving the Optimization Problem The optimization problem is solved using:
When needed, the objective function Hessian can be defined using a LinearOperator object,
or a Hessian-vector product through the parameter hessp .
Alternatively, the first and second derivatives of the objective function can be approximated. For instance, the Hessian can be approximated with SR1 quasi-Newton approximation and the gradient with finite differences.
Byrd, Richard H., Mary E. Hribar, and Jorge Nocedal. 1999. An interior point algorithm for large-scale nonlinear programming. SIAM Journal on Optimization 9.4: 877-900.
Lalee, Marucha, Jorge Nocedal, and Todd Plantega. 1998. On the implementation of an algorithm for large-scale equality constrained optimization. SIAM Journal on Optimization 8.3: 682-706.
Sequential Least SQuares Programming (SLSQP) Algorithm ( method='SLSQP' ) #
The SLSQP method deals with constrained minimization problems of the form:
Where \(\mathcal{E}\) or \(\mathcal{I}\) are sets of indices containing equality and inequality constraints.
Both linear and nonlinear constraints are defined as dictionaries with keys type , fun and jac .
And the optimization problem is solved with:
Most of the options available for the method 'trust-constr' are not available for 'SLSQP' .
Local minimization solver comparison #
Find a solver that meets your requirements using the table below. If there are multiple candidates, try several and see which ones best meet your needs (e.g. execution time, objective function value).
Solver | Bounds Constraints | Nonlinear Constraints | Uses Gradient | Uses Hessian | Utilizes Sparsity |
|---|---|---|---|---|---|
CG | ✓ | ||||
BFGS | ✓ | ||||
dogleg | ✓ | ✓ | |||
trust-ncg | ✓ | ✓ | |||
trust-krylov | ✓ | ✓ | |||
trust-exact | ✓ | ✓ | |||
Newton-CG | ✓ | ✓ | ✓ | ||
Nelder-Mead | ✓ | ||||
Powell | ✓ | ||||
L-BFGS-B | ✓ | ✓ | |||
TNC | ✓ | ✓ | |||
COBYLA | ✓ | ✓ | |||
SLSQP | ✓ | ✓ | ✓ | ||
trust-constr | ✓ | ✓ | ✓ | ✓ | ✓ |
Global optimization #
Global optimization aims to find the global minimum of a function within given bounds, in the presence of potentially many local minima. Typically, global minimizers efficiently search the parameter space, while using a local minimizer (e.g., minimize ) under the hood. SciPy contains a number of good global optimizers. Here, we’ll use those on the same objective function, namely the (aptly named) eggholder function:
This function looks like an egg carton:
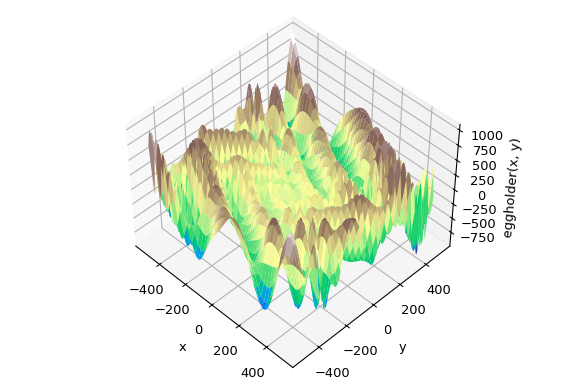
We now use the global optimizers to obtain the minimum and the function value at the minimum. We’ll store the results in a dictionary so we can compare different optimization results later.
All optimizers return an OptimizeResult , which in addition to the solution contains information on the number of function evaluations, whether the optimization was successful, and more. For brevity, we won’t show the full output of the other optimizers:
shgo has a second method, which returns all local minima rather than only what it thinks is the global minimum:
We’ll now plot all found minima on a heatmap of the function:
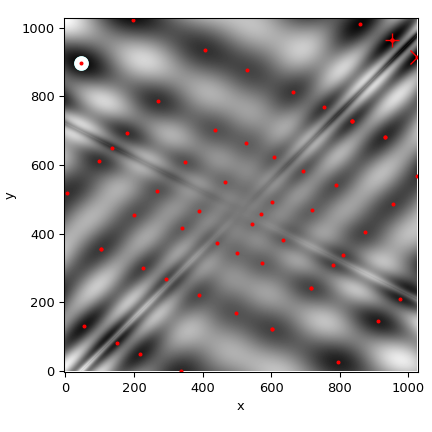
Comparison of Global Optimizers #
Solver | Bounds Constraints | Nonlinear Constraints | Uses Gradient | Uses Hessian |
|---|---|---|---|---|
basinhopping | (✓) | (✓) | ||
direct | ✓ | |||
dual_annealing | ✓ | (✓) | (✓) | |
differential_evolution | ✓ | ✓ | ||
shgo | ✓ | ✓ | (✓) | (✓) |
(✓) = Depending on the chosen local minimizer
Least-squares minimization ( least_squares ) #
SciPy is capable of solving robustified bound-constrained nonlinear least-squares problems:
Here \(f_i(\mathbf{x})\) are smooth functions from \(\mathbb{R}^n\) to \(\mathbb{R}\) , we refer to them as residuals. The purpose of a scalar-valued function \(\rho(\cdot)\) is to reduce the influence of outlier residuals and contribute to robustness of the solution, we refer to it as a loss function. A linear loss function gives a standard least-squares problem. Additionally, constraints in a form of lower and upper bounds on some of \(x_j\) are allowed.
All methods specific to least-squares minimization utilize a \(m \times n\) matrix of partial derivatives called Jacobian and defined as \(J_{ij} = \partial f_i / \partial x_j\) . It is highly recommended to compute this matrix analytically and pass it to least_squares , otherwise, it will be estimated by finite differences, which takes a lot of additional time and can be very inaccurate in hard cases.
Function least_squares can be used for fitting a function \(\varphi(t; \mathbf{x})\) to empirical data \(\{(t_i, y_i), i = 0, \ldots, m-1\}\) . To do this, one should simply precompute residuals as \(f_i(\mathbf{x}) = w_i (\varphi(t_i; \mathbf{x}) - y_i)\) , where \(w_i\) are weights assigned to each observation.
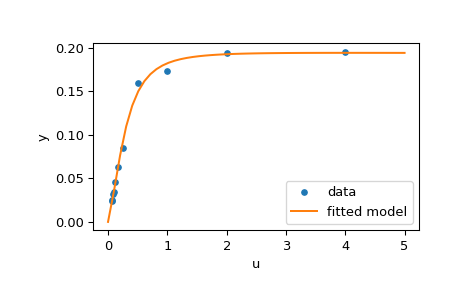
Example of solving a fitting problem #
Here we consider an enzymatic reaction [ 1 ] . There are 11 residuals defined as
where \(y_i\) are measurement values and \(u_i\) are values of the independent variable. The unknown vector of parameters is \(\mathbf{x} = (x_0, x_1, x_2, x_3)^T\) . As was said previously, it is recommended to compute Jacobian matrix in a closed form:
We are going to use the “hard” starting point defined in [ 2 ] . To find a physically meaningful solution, avoid potential division by zero and assure convergence to the global minimum we impose constraints \(0 \leq x_j \leq 100, j = 0, 1, 2, 3\) .
The code below implements least-squares estimation of \(\mathbf{x}\) and finally plots the original data and the fitted model function:
Further examples #
Three interactive examples below illustrate usage of least_squares in greater detail.
Large-scale bundle adjustment in scipy demonstrates large-scale capabilities of least_squares and how to efficiently compute finite difference approximation of sparse Jacobian.
Robust nonlinear regression in scipy shows how to handle outliers with a robust loss function in a nonlinear regression.
Solving a discrete boundary-value problem in scipy examines how to solve a large system of equations and use bounds to achieve desired properties of the solution.
For the details about mathematical algorithms behind the implementation refer to documentation of least_squares .
Univariate function minimizers ( minimize_scalar ) #
Often only the minimum of an univariate function (i.e., a function that takes a scalar as input) is needed. In these circumstances, other optimization techniques have been developed that can work faster. These are accessible from the minimize_scalar function, which proposes several algorithms.
Unconstrained minimization ( method='brent' ) #
There are, actually, two methods that can be used to minimize an univariate function: brent and golden , but golden is included only for academic purposes and should rarely be used. These can be respectively selected through the method parameter in minimize_scalar . The brent method uses Brent’s algorithm for locating a minimum. Optimally, a bracket (the bracket parameter) should be given which contains the minimum desired. A bracket is a triple \(\left( a, b, c \right)\) such that \(f \left( a \right) > f \left( b \right) < f \left( c \right)\) and \(a < b < c\) . If this is not given, then alternatively two starting points can be chosen and a bracket will be found from these points using a simple marching algorithm. If these two starting points are not provided, 0 and 1 will be used (this may not be the right choice for your function and result in an unexpected minimum being returned).
Here is an example:
Bounded minimization ( method='bounded' ) #
Very often, there are constraints that can be placed on the solution space before minimization occurs. The bounded method in minimize_scalar is an example of a constrained minimization procedure that provides a rudimentary interval constraint for scalar functions. The interval constraint allows the minimization to occur only between two fixed endpoints, specified using the mandatory bounds parameter.
For example, to find the minimum of \(J_{1}\left( x \right)\) near \(x=5\) , minimize_scalar can be called using the interval \(\left[ 4, 7 \right]\) as a constraint. The result is \(x_{\textrm{min}}=5.3314\) :
Custom minimizers #
Sometimes, it may be useful to use a custom method as a (multivariate or univariate) minimizer, for example, when using some library wrappers of minimize (e.g., basinhopping ).
We can achieve that by, instead of passing a method name, passing a callable (either a function or an object implementing a __call__ method) as the method parameter.
Let us consider an (admittedly rather virtual) need to use a trivial custom multivariate minimization method that will just search the neighborhood in each dimension independently with a fixed step size:
This will work just as well in case of univariate optimization:
Root finding #
Scalar functions #.
If one has a single-variable equation, there are multiple different root finding algorithms that can be tried. Most of these algorithms require the endpoints of an interval in which a root is expected (because the function changes signs). In general, brentq is the best choice, but the other methods may be useful in certain circumstances or for academic purposes. When a bracket is not available, but one or more derivatives are available, then newton (or halley , secant ) may be applicable. This is especially the case if the function is defined on a subset of the complex plane, and the bracketing methods cannot be used.
Fixed-point solving #
A problem closely related to finding the zeros of a function is the problem of finding a fixed point of a function. A fixed point of a function is the point at which evaluation of the function returns the point: \(g\left(x\right)=x.\) Clearly, the fixed point of \(g\) is the root of \(f\left(x\right)=g\left(x\right)-x.\) Equivalently, the root of \(f\) is the fixed point of \(g\left(x\right)=f\left(x\right)+x.\) The routine fixed_point provides a simple iterative method using Aitkens sequence acceleration to estimate the fixed point of \(g\) given a starting point.
Sets of equations #
Finding a root of a set of non-linear equations can be achieved using the root function. Several methods are available, amongst which hybr (the default) and lm , which, respectively, use the hybrid method of Powell and the Levenberg-Marquardt method from MINPACK.
The following example considers the single-variable transcendental equation
a root of which can be found as follows:
Consider now a set of non-linear equations
We define the objective function so that it also returns the Jacobian and indicate this by setting the jac parameter to True . Also, the Levenberg-Marquardt solver is used here.

Root finding for large problems #
Methods hybr and lm in root cannot deal with a very large number of variables ( N ), as they need to calculate and invert a dense N x N Jacobian matrix on every Newton step. This becomes rather inefficient when N grows.
Consider, for instance, the following problem: we need to solve the following integrodifferential equation on the square \([0,1]\times[0,1]\) :
with the boundary condition \(P(x,1) = 1\) on the upper edge and \(P=0\) elsewhere on the boundary of the square. This can be done by approximating the continuous function P by its values on a grid, \(P_{n,m}\approx{}P(n h, m h)\) , with a small grid spacing h . The derivatives and integrals can then be approximated; for instance \(\partial_x^2 P(x,y)\approx{}(P(x+h,y) - 2 P(x,y) + P(x-h,y))/h^2\) . The problem is then equivalent to finding the root of some function residual(P) , where P is a vector of length \(N_x N_y\) .
Now, because \(N_x N_y\) can be large, methods hybr or lm in root will take a long time to solve this problem. The solution can, however, be found using one of the large-scale solvers, for example krylov , broyden2 , or anderson . These use what is known as the inexact Newton method, which instead of computing the Jacobian matrix exactly, forms an approximation for it.
The problem we have can now be solved as follows:
Still too slow? Preconditioning. #
When looking for the zero of the functions \(f_i({\bf x}) = 0\) , i = 1, 2, …, N , the krylov solver spends most of the time inverting the Jacobian matrix,
If you have an approximation for the inverse matrix \(M\approx{}J^{-1}\) , you can use it for preconditioning the linear-inversion problem. The idea is that instead of solving \(J{\bf s}={\bf y}\) one solves \(MJ{\bf s}=M{\bf y}\) : since matrix \(MJ\) is “closer” to the identity matrix than \(J\) is, the equation should be easier for the Krylov method to deal with.
The matrix M can be passed to root with method krylov as an option options['jac_options']['inner_M'] . It can be a (sparse) matrix or a scipy.sparse.linalg.LinearOperator instance.
For the problem in the previous section, we note that the function to solve consists of two parts: the first one is the application of the Laplace operator, \([\partial_x^2 + \partial_y^2] P\) , and the second is the integral. We can actually easily compute the Jacobian corresponding to the Laplace operator part: we know that in 1-D
so that the whole 2-D operator is represented by
The matrix \(J_2\) of the Jacobian corresponding to the integral is more difficult to calculate, and since all of it entries are nonzero, it will be difficult to invert. \(J_1\) on the other hand is a relatively simple matrix, and can be inverted by scipy.sparse.linalg.splu (or the inverse can be approximated by scipy.sparse.linalg.spilu ). So we are content to take \(M\approx{}J_1^{-1}\) and hope for the best.
In the example below, we use the preconditioner \(M=J_1^{-1}\) .
Resulting run, first without preconditioning:
and then with preconditioning:
Using a preconditioner reduced the number of evaluations of the residual function by a factor of 4 . For problems where the residual is expensive to compute, good preconditioning can be crucial — it can even decide whether the problem is solvable in practice or not.
Preconditioning is an art, science, and industry. Here, we were lucky in making a simple choice that worked reasonably well, but there is a lot more depth to this topic than is shown here.
Linear programming ( linprog ) #
The function linprog can minimize a linear objective function subject to linear equality and inequality constraints. This kind of problem is well known as linear programming. Linear programming solves problems of the following form:
where \(x\) is a vector of decision variables; \(c\) , \(b_{ub}\) , \(b_{eq}\) , \(l\) , and \(u\) are vectors; and \(A_{ub}\) and \(A_{eq}\) are matrices.
In this tutorial, we will try to solve a typical linear programming problem using linprog .
Linear programming example #
Consider the following simple linear programming problem:
We need some mathematical manipulations to convert the target problem to the form accepted by linprog .
First of all, let’s consider the objective function. We want to maximize the objective function, but linprog can only accept a minimization problem. This is easily remedied by converting the maximize \(29x_1 + 45x_2\) to minimizing \(-29x_1 -45x_2\) . Also, \(x_3, x_4\) are not shown in the objective function. That means the weights corresponding with \(x_3, x_4\) are zero. So, the objective function can be converted to:
If we define the vector of decision variables \(x = [x_1, x_2, x_3, x_4]^T\) , the objective weights vector \(c\) of linprog in this problem should be
Next, let’s consider the two inequality constraints. The first one is a “less than” inequality, so it is already in the form accepted by linprog . The second one is a “greater than” inequality, so we need to multiply both sides by \(-1\) to convert it to a “less than” inequality. Explicitly showing zero coefficients, we have:
These equations can be converted to matrix form:
Next, let’s consider the two equality constraints. Showing zero weights explicitly, these are:
Lastly, let’s consider the separate inequality constraints on individual decision variables, which are known as “box constraints” or “simple bounds”. These constraints can be applied using the bounds argument of linprog . As noted in the linprog documentation, the default value of bounds is (0, None) , meaning that the lower bound on each decision variable is 0, and the upper bound on each decision variable is infinity: all the decision variables are non-negative. Our bounds are different, so we will need to specify the lower and upper bound on each decision variable as a tuple and group these tuples into a list.
Finally, we can solve the transformed problem using linprog .
The result states that our problem is infeasible, meaning that there is no solution vector that satisfies all the constraints. That doesn’t necessarily mean we did anything wrong; some problems truly are infeasible. Suppose, however, that we were to decide that our bound constraint on \(x_1\) was too tight and that it could be loosened to \(0 \leq x_1 \leq 6\) . After adjusting our code x1_bounds = (0, 6) to reflect the change and executing it again:
The result shows the optimization was successful. We can check the objective value ( result.fun ) is same as \(c^Tx\) :
We can also check that all constraints are satisfied within reasonable tolerances:
Assignment problems #
Linear sum assignment problem example #.
Consider the problem of selecting students for a swimming medley relay team. We have a table showing times for each swimming style of five students:
Student | backstroke | breaststroke | butterfly | freestyle |
|---|---|---|---|---|
A | 43.5 | 47.1 | 48.4 | 38.2 |
B | 45.5 | 42.1 | 49.6 | 36.8 |
C | 43.4 | 39.1 | 42.1 | 43.2 |
D | 46.5 | 44.1 | 44.5 | 41.2 |
E | 46.3 | 47.8 | 50.4 | 37.2 |
We need to choose a student for each of the four swimming styles such that the total relay time is minimized. This is a typical linear sum assignment problem. We can use linear_sum_assignment to solve it.
The linear sum assignment problem is one of the most famous combinatorial optimization problems. Given a “cost matrix” \(C\) , the problem is to choose
exactly one element from each row
without choosing more than one element from any column
such that the sum of the chosen elements is minimized
In other words, we need to assign each row to one column such that the sum of the corresponding entries is minimized.
Formally, let \(X\) be a boolean matrix where \(X[i,j] = 1\) iff row \(i\) is assigned to column \(j\) . Then the optimal assignment has cost
The first step is to define the cost matrix. In this example, we want to assign each swimming style to a student. linear_sum_assignment is able to assign each row of a cost matrix to a column. Therefore, to form the cost matrix, the table above needs to be transposed so that the rows correspond with swimming styles and the columns correspond with students:
We can solve the assignment problem with linear_sum_assignment :
The row_ind and col_ind are optimal assigned matrix indexes of the cost matrix:
The optimal assignment is:
The optimal total medley time is:
Note that this result is not the same as the sum of the minimum times for each swimming style:
because student “C” is the best swimmer in both “breaststroke” and “butterfly” style. We cannot assign student “C” to both styles, so we assigned student C to the “breaststroke” style and D to the “butterfly” style to minimize the total time.
Some further reading and related software, such as Newton-Krylov [KK] , PETSc [PP] , and PyAMG [AMG] :
D.A. Knoll and D.E. Keyes, “Jacobian-free Newton-Krylov methods”, J. Comp. Phys. 193, 357 (2004). DOI:10.1016/j.jcp.2003.08.010
PETSc https://www.mcs.anl.gov/petsc/ and its Python bindings https://bitbucket.org/petsc/petsc4py/
PyAMG (algebraic multigrid preconditioners/solvers) pyamg/pyamg#issues
Mixed integer linear programming #
Knapsack problem example #.
The knapsack problem is a well known combinatorial optimization problem. Given a set of items, each with a size and a value, the problem is to choose the items that maximize the total value under the condition that the total size is below a certain threshold.
Formally, let
\(x_i\) be a boolean variable that indicates whether item \(i\) is included in the knapsack,
\(n\) be the total number of items,
\(v_i\) be the value of item \(i\) ,
\(s_i\) be the size of item \(i\) , and
\(C\) be the capacity of the knapsack.
Then the problem is:
Although the objective function and inequality constraints are linear in the decision variables \(x_i\) , this differs from a typical linear programming problem in that the decision variables can only assume integer values. Specifically, our decision variables can only be \(0\) or \(1\) , so this is known as a binary integer linear program (BILP). Such a problem falls within the larger class of mixed integer linear programs (MILPs), which we we can solve with milp .
In our example, there are 8 items to choose from, and the size and value of each is specified as follows.
We need to constrain our eight decision variables to be binary. We do so by adding a Bounds : constraint to ensure that they lie between \(0\) and \(1\) , and we apply “integrality” constraints to ensure that they are either \(0\) or \(1\) .
The knapsack capacity constraint is specified using LinearConstraint .
If we are following the usual rules of linear algebra, the input A should be a two-dimensional matrix, and the lower and upper bounds lb and ub should be one-dimensional vectors, but LinearConstraint is forgiving as long as the inputs can be broadcast to consistent shapes.
Using the variables defined above, we can solve the knapsack problem using milp . Note that milp minimizes the objective function, but we want to maximize the total value, so we set c to be negative of the values.
Let’s check the result:
This means that we should select the items 1, 2, 4, 5, 6 to optimize the total value under the size constraint. Note that this is different from we would have obtained had we solved the linear programming relaxation (without integrality constraints) and attempted to round the decision variables.
If we were to round this solution up to array([1., 1., 1., 1., 1., 1., 0., 0.]) , our knapsack would be over the capacity constraint, whereas if we were to round down to array([1., 1., 1., 1., 0., 1., 0., 0.]) , we would have a sub-optimal solution.
For more MILP tutorials, see the Jupyter notebooks on SciPy Cookbooks:
Compressed Sensing l1 program
Compressed Sensing l0 program
Instantly share code, notes, and snippets.
wenxin-liu / Optimization Using Gradient Descent: Linear Regression.ipynb
- Download ZIP
- Star ( 0 ) 0 You must be signed in to star a gist
- Fork ( 0 ) 0 You must be signed in to fork a gist
- Embed Embed this gist in your website.
- Share Copy sharable link for this gist.
- Clone via HTTPS Clone using the web URL.
- Learn more about clone URLs
- Save wenxin-liu/ca51a97500833af5985f19a543fe62b2 to your computer and use it in GitHub Desktop.
Deep-Learning-Specialization
Coursera deep learning specialization, neural networks and deep learning.
In this course, you will learn the foundations of deep learning. When you finish this class, you will:
- Understand the major technology trends driving Deep Learning.
- Be able to build, train and apply fully connected deep neural networks.
- Know how to implement efficient (vectorized) neural networks.
- Understand the key parameters in a neural network’s architecture.
Week 1: Introduction to deep learning
Be able to explain the major trends driving the rise of deep learning, and understand where and how it is applied today.
- Quiz 1: Introduction to deep learning
Week 2: Neural Networks Basics
Learn to set up a machine learning problem with a neural network mindset. Learn to use vectorization to speed up your models.
- Quiz 2: Neural Network Basics
- Programming Assignment: Python Basics With Numpy
- Programming Assignment: Logistic Regression with a Neural Network mindset
Week 3: Shallow neural networks
Learn to build a neural network with one hidden layer, using forward propagation and backpropagation.
- Quiz 3: Shallow Neural Networks
- Programming Assignment: Planar Data Classification with Onehidden Layer
Week 4: Deep Neural Networks
Understand the key computations underlying deep learning, use them to build and train deep neural networks, and apply it to computer vision.
- Quiz 4: Key concepts on Deep Neural Networks
- Programming Assignment: Building your Deep Neural Network Step by Step
- Programming Assignment: Deep Neural Network Application
Course Certificate
Optimization Methods
These are my personal programming assignments at the 2nd week after studying the course Improving Deep Neural Networks: Hyperparameter tuning, Regularization and Optimization and the copyright belongs to deeplearning.ai .
Until now, you’ve always used Gradient Descent to update the parameters and minimize the cost. In this notebook, you will learn more advanced optimization methods that can speed up learning and perhaps even get you to a better final value for the cost function. Having a good optimization algorithm can be the difference between waiting days vs. just a few hours to get a good result.
Gradient descent goes “downhill” on a cost function $J$. Think of it as trying to do this:
Notations : As usual, $\frac{∂J}{∂a}= da$ for any variable $a$.
To get started, run the following code to import the libraries you will need.
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 | numpy as np import matplotlib.pyplot as plt import scipy.io import math import sklearn import sklearn.datasets from opt_utils import load_params_and_grads, initialize_parameters, forward_propagation, backward_propagation from opt_utils import compute_cost, predict, predict_dec, plot_decision_boundary, load_dataset from testCases import * %matplotlib inline plt.rcParams['figure.figsize'] = (7.0, 4.0) # set default size of plots plt.rcParams['image.interpolation'] = 'nearest' plt.rcParams['image.cmap'] = 'gray' |
1. Gradient Descent
A simple optimization method in machine learning is gradient descent (GD). When you take gradient steps with respect to all $m$ examples on each step, it is also called Batch Gradient Descent.
Warm-up exercise: Implement the gradient descent update rule. The gradient descent rule is, for $l=1,…,L$: $$W^{[l]} = W^{[l]} - \alpha \text{ } dW^{[l]} \tag{1}$$ $$b^{[l]} = b^{[l]} - \alpha \text{ } db^{[l]} \tag{2}$$
where $L$ is the number of layers and $α$ is the learning rate. All parameters should be stored in the parameters dictionary.
Note that the iterator $l$ starts at $0$ in the for loop while the first parameters are $W^{[1]}$ and $b^{[1]}$. You need to shift $l$ to $l+1$ when coding.
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 | update_parameters_with_gd(parameters, grads, learning_rate): """ Update parameters using one step of gradient descent Arguments: parameters -- python dictionary containing your parameters to be updated: parameters['W' + str(l)] = Wl parameters['b' + str(l)] = bl grads -- python dictionary containing your gradients to update each parameters: grads['dW' + str(l)] = dWl grads['db' + str(l)] = dbl learning_rate -- the learning rate, scalar. Returns: parameters -- python dictionary containing your updated parameters """ L = len(parameters) // 2; # number of layers in the neural networks # Update rule for each parameter for l in range(L): ### START CODE HERE ### (approx. 2 lines) parameters['W' + str(l + 1)] -= learning_rate * grads['dW' + str(l + 1)]; parameters['b' + str(l + 1)] -= learning_rate * grads['db' + str(l + 1)]; ### END CODE HERE ### return parameters; |
| 2 3 4 5 6 7 | parameters = update_parameters_with_gd(parameters, grads, learning_rate); print("W1 = " + str(parameters["W1"])); print("b1 = " + str(parameters["b1"])); print("W2 = " + str(parameters["W2"])); print("b2 = " + str(parameters["b2"])); |
Expected Output :
| value | |
|---|---|
| [[ 1.63535156 -0.62320365 -0.53718766] [-1.07799357 0.85639907 -2.29470142]] | |
| [[ 1.74604067] [-0.75184921]] | |
| [[ 0.32171798 -0.25467393 1.46902454] [-2.05617317 -0.31554548 -0.3756023 ] [ 1.1404819 -1.09976462 -0.1612551 ]] | |
| [[-0.88020257] [ 0.02561572] [ 0.57539477]] |
A variant of this is Stochastic Gradient Descent (SGD), which is equivalent to mini-batch gradient descent where each mini-batch has just 1 example. The update rule that you have just implemented does not change. What changes is that you would be computing gradients on just one training example at a time, rather than on the whole training set. The code examples below illustrate the difference between stochastic gradient descent and (batch) gradient descent.
(Batch) Gradient Descent :
| 2 3 4 5 6 7 8 9 10 11 12 | Y = labels parameters = initialize_parameters(layers_dims) for i in range(0, num_iterations): # Forward propagation a, caches = forward_propagation(X, parameters) # Compute cost. cost = compute_cost(a, Y) # Backward propagation. grads = backward_propagation(a, caches, parameters) # Update parameters. parameters = update_parameters(parameters, grads) |
Stochastic Gradient Descent :
| 2 3 4 5 6 7 8 9 10 11 12 13 | Y = labels parameters = initialize_parameters(layers_dims) for i in range(0, num_iterations): for j in range(0, m): # Forward propagation a, caches = forward_propagation(X[:,j], parameters) # Compute cost cost = compute_cost(a, Y[:,j]) # Backward propagation grads = backward_propagation(a, caches, parameters) # Update parameters. parameters = update_parameters(parameters, grads) |
In Stochastic Gradient Descent, you use only 1 training example before updating the gradients. When the training set is large, SGD can be faster. But the parameters will “oscillate” toward the minimum rather than converge smoothly. Here is an illustration of this:
Note also that implementing SGD requires 3 for-loops in total:
- Over the number of iterations
- Over the m training examples
- Over the layers (to update all parameters, from ($W^{[1]}$,$b^{[1]}$) to ($W^{[L]}$,$b^{[L]}$)
In practice, you’ll often get faster results if you do not use neither the whole training set, nor only one training example, to perform each update. Mini-batch gradient descent uses an intermediate number of examples for each step. With mini-batch gradient descent, you loop over the mini-batches instead of looping over individual training examples.
What you should remember:
- The difference between gradient descent, mini-batch gradient descent and stochastic gradient descent is the number of examples you use to perform one update step.
- You have to tune a learning rate hyperparameter α.
- With a well-turned mini-batch size, usually it outperforms either gradient descent or stochastic gradient descent (particularly when the training set is large).
2. Mini-Batch Gradient descent
Let’s learn how to build mini-batches from the training set $(X, Y)$.
There are two steps:
- Shuffle : Create a shuffled version of the training set $(X, Y)$ as shown below. Each column of $X$ and $Y$ represents a training example. Note that the random shuffling is done synchronously between $X$ and $Y$. Such that after the shuffling the ith column of $X$ is the example corresponding to the ith label in $Y$. The shuffling step ensures that examples will be split randomly into different mini-batches.
- Partition : Partition the shuffled $(X, Y)$ into mini-batches of size mini_batch_size (here 64). Note that the number of training examples is not always divisible by mini_batch_size . The last mini batch might be smaller, but you don’t need to worry about this. When the final mini-batch is smaller than the full mini_batch_size , it will look like this:
Exercise : Implement random_mini_batches . We coded the shuffling part for you. To help you with the partitioning step, we give you the following code that selects the indexes for the 1st and 2nd mini-batches:
| 2 3 | : mini_batch_size] second_mini_batch_X = shuffled_X[:, mini_batch_size : 2 * mini_batch_size] ... |
Note that the last mini-batch might end up smaller than mini_batch_size=64 . Let $\lfloor s \rfloor$ represents $s$ rounded down to the nearest integer (this is math.floor(s) in Python). If the total number of examples is not a multiple of mini_batch_size=64 then there will be $\lfloor \frac{m}{mini_batch_size}\rfloor$ mini-batches with a full 64 examples, and the number of examples in the final mini-batch will be $(m-mini__batch__size \times \lfloor \frac{m}{mini_batch_size}\rfloor)$.
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 | def random_mini_batches(X, Y, mini_batch_size = 64, seed = 0): """ Creates a list of random minibatches from (X, Y) Arguments: X -- input data, of shape (input size, number of examples) Y -- true "label" vector (1 for blue dot / 0 for red dot), of shape (1, number of examples) mini_batch_size -- size of the mini-batches, integer Returns: mini_batches -- list of synchronous (mini_batch_X, mini_batch_Y) """ np.random.seed(seed) # To make your "random" minibatches the same as ours m = X.shape[1] # number of training examples mini_batches = [] # Step 1: Shuffle (X, Y) permutation = list(np.random.permutation(m)) shuffled_X = X[:, permutation] shuffled_Y = Y[:, permutation].reshape((1,m)) # Step 2: Partition (shuffled_X, shuffled_Y). Minus the end case. num_complete_minibatches = math.floor(m/mini_batch_size) # number of mini batches of size mini_batch_size in your partitionning for k in range(0, num_complete_minibatches): ### START CODE HERE ### (approx. 2 lines) mini_batch_X = shuffled_X[:, k * mini_batch_size : (k + 1) * mini_batch_size]; mini_batch_Y = shuffled_Y[:, k * mini_batch_size : (k + 1) * mini_batch_size]; ### END CODE HERE ### mini_batch = (mini_batch_X, mini_batch_Y) mini_batches.append(mini_batch) # Handling the end case (last mini-batch < mini_batch_size) if m % mini_batch_size != 0: ### START CODE HERE ### (approx. 2 lines) mini_batch_X = shuffled_X[:, mini_batch_size * num_complete_minibatches : m]; mini_batch_Y = shuffled_Y[:, mini_batch_size * num_complete_minibatches : m]; ### END CODE HERE ### mini_batch = (mini_batch_X, mini_batch_Y) mini_batches.append(mini_batch) return mini_batches |
| 2 3 4 5 6 7 8 9 10 | mini_batches = random_mini_batches(X_assess, Y_assess, mini_batch_size) print ("shape of the 1st mini_batch_X: " + str(mini_batches[0][0].shape)) print ("shape of the 2nd mini_batch_X: " + str(mini_batches[1][0].shape)) print ("shape of the 3rd mini_batch_X: " + str(mini_batches[2][0].shape)) print ("shape of the 1st mini_batch_Y: " + str(mini_batches[0][1].shape)) print ("shape of the 2nd mini_batch_Y: " + str(mini_batches[1][1].shape)) print ("shape of the 3rd mini_batch_Y: " + str(mini_batches[2][1].shape)) print ("mini batch sanity check: " + str(mini_batches[0][0][0][0:3])) |
| value | |
|---|---|
| (12288, 64) | |
| (12288, 64) | |
| (12288, 20) | |
| (1, 64) | |
| (1, 64) | |
| (1, 20) | |
| [ 0.90085595 -0.7612069 0.2344157 ] |
What you should remember :
- Shuffling and Partitioning are the two steps required to build mini-batches
- Powers of two are often chosen to be the mini-batch size, e.g., 16, 32, 64, 128.
3. Momentum
Because mini-batch gradient descent makes a parameter update after seeing just a subset of examples, the direction of the update has some variance, and so the path taken by mini-batch gradient descent will “oscillate” toward convergence. Using momentum can reduce these oscillations.
Momentum takes into account the past gradients to smooth out the update. We will store the ‘direction’ of the previous gradients in the variable $v$. Formally, this will be the exponentially weighted average of the gradient on previous steps. You can also think of $v$ as the “velocity” of a ball rolling downhill, building up speed (and momentum) according to the direction of the gradient/slope of the hill.
Exercise : Initialize the velocity. The velocity, $v$, is a python dictionary that needs to be initialized with arrays of zeros. Its keys are the same as those in the grads dictionary, that is:
| 2 3 | l=1,...,L: v["dW" + str(l+1)] = ... #(numpy array of zeros with the same shape as parameters["W" + str(l+1)]) v["db" + str(l+1)] = ... #(numpy array of zeros with the same shape as parameters["b" + str(l+1)]) |
Note that the iterator $l$ starts at $0$ in the for loop while the first parameters are v[“dW1”] and v[“db1”] (that’s a “one” on the superscript). This is why we are shifting l to l + 1 in the for loop.
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 | def initialize_velocity(parameters): """ Initializes the velocity as a python dictionary with: - keys: "dW1", "db1", ..., "dWL", "dbL" - values: numpy arrays of zeros of the same shape as the corresponding gradients/parameters. Arguments: parameters -- python dictionary containing your parameters. parameters['W' + str(l)] = Wl parameters['b' + str(l)] = bl Returns: v -- python dictionary containing the current velocity. v['dW' + str(l)] = velocity of dWl v['db' + str(l)] = velocity of dbl """ L = len(parameters) // 2 # number of layers in the neural networks v = {} # Initialize velocity for l in range(L): ### START CODE HERE ### (approx. 2 lines) v['dW' + str(l + 1)] = np.zeros(parameters['W' + str(l + 1)].shape); v['db' + str(l + 1)] = np.zeros(parameters['b' + str(l + 1)].shape); ### END CODE HERE ### return v |
| 2 3 4 5 6 7 | v = initialize_velocity(parameters) print("v[\"dW1\"] = " + str(v["dW1"])) print("v[\"db1\"] = " + str(v["db1"])) print("v[\"dW2\"] = " + str(v["dW2"])) print("v[\"db2\"] = " + str(v["db2"])) |
| value | |
|---|---|
| [[ 0. 0. 0.] [ 0. 0. 0.]] | |
| [[ 0.] [ 0.]] | |
| [[ 0. 0. 0.] [ 0. 0. 0.] [ 0. 0. 0.]] | |
| [[ 0.] [ 0.] [ 0.]] |
Exercise : Now, implement the parameters update with momentum. The momentum update rule is, for l=1,...,L : $$ \begin{cases} v_{dW^{[l]}} = \beta v_{dW^{[l]}} + (1 - \beta) dW^{[l]} \ W^{[l]} = W^{[l]} - \alpha v_{dW^{[l]}} \end{cases}\tag{3} $$ $$ \begin{cases} v_{db^{[l]}} = \beta v_{db^{[l]}} + (1 - \beta) db^{[l]} \ b^{[l]} = b^{[l]} - \alpha v_{db^{[l]}} \end{cases}\tag{4} $$
where $L$ is the number of layers, $β$ is the momentum and $α$ is the learning rate. All parameters should be stored in the parameters dictionary. Note that the iterator $l$ starts at $0$ in the for loop while the first parameters are $W^{[1]}$ and $b^{[1]}$ (that’s a “one” on the superscript). So you will need to shift $l$ to $l + 1$ when coding.
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 | def update_parameters_with_momentum(parameters, grads, v, beta, learning_rate): """ Update parameters using Momentum Arguments: parameters -- python dictionary containing your parameters: parameters['W' + str(l)] = Wl parameters['b' + str(l)] = bl grads -- python dictionary containing your gradients for each parameters: grads['dW' + str(l)] = dWl grads['db' + str(l)] = dbl v -- python dictionary containing the current velocity: v['dW' + str(l)] = ... v['db' + str(l)] = ... beta -- the momentum hyperparameter, scalar learning_rate -- the learning rate, scalar Returns: parameters -- python dictionary containing your updated parameters v -- python dictionary containing your updated velocities """ L = len(parameters) // 2 # number of layers in the neural networks # Momentum update for each parameter for l in range(L): ### START CODE HERE ### (approx. 4 lines) # compute velocities v['dW' + str(l + 1)] = beta * v['dW' + str(l + 1)] + (1 - beta) * grads['dW' + str(l + 1)]; v['db' + str(l + 1)] = beta * v['db' + str(l + 1)] + (1 - beta) * grads['db' + str(l + 1)]; # update parameters parameters['W' + str(l + 1)] -= learning_rate * v['dW' + str(l + 1)]; parameters['b' + str(l + 1)] -= learning_rate * v['db' + str(l + 1)]; ### END CODE HERE ### return parameters, v |
| 2 3 4 5 6 7 8 9 10 11 | parameters, v = update_parameters_with_momentum(parameters, grads, v, beta = 0.9, learning_rate = 0.01) print("W1 = " + str(parameters["W1"])) print("b1 = " + str(parameters["b1"])) print("W2 = " + str(parameters["W2"])) print("b2 = " + str(parameters["b2"])) print("v[\"dW1\"] = " + str(v["dW1"])) print("v[\"db1\"] = " + str(v["db1"])) print("v[\"dW2\"] = " + str(v["dW2"])) print("v[\"db2\"] = " + str(v["db2"])) |
| value | |
|---|---|
| [[ 1.62544598 -0.61290114 -0.52907334] [-1.07347112 0.86450677 -2.30085497]] | |
| [[ 1.74493465] [-0.76027113]] | |
| [[ 0.31930698 -0.24990073 1.4627996 ] [-2.05974396 -0.32173003 -0.38320915] [ 1.13444069 -1.0998786 -0.1713109 ]] | |
| [[-0.87809283] [ 0.04055394] [ 0.58207317]] | |
| [[-0.11006192 0.11447237 0.09015907] [ 0.05024943 0.09008559 -0.06837279]] | |
| [[-0.01228902] [-0.09357694]] | |
| [[-0.02678881 0.05303555 -0.06916608] [-0.03967535 -0.06871727 -0.08452056] [-0.06712461 -0.00126646 -0.11173103]] | |
| [[ 0.02344157][ 0.16598022] [ 0.07420442]] |
- The velocity is initialized with zeros. So the algorithm will take a few iterations to “build up” velocity and start to take bigger steps.
- If $β=0$, then this just becomes standard gradient descent without momentum.
How do you choose $β$ ?
The larger the momentum $β$ is, the smoother the update because the more we take the past gradients into account. But if $β$ is too big, it could also smooth out the updates too much. Common values for $β$ range from 0.8 to 0.999. If you don’t feel inclined to tune this, $β=0.9$ is often a reasonable default. Tuning the optimal $β$ for your model might need trying several values to see what works best in term of reducing the value of the cost function $J$.
- Momentum takes past gradients into account to smooth out the steps of gradient descent. It can be applied with batch gradient descent, mini-batch gradient descent or stochastic gradient descent.
- You have to tune a momentum hyperparameter $β$ and a learning rate $α$.
Adam is one of the most effective optimization algorithms for training neural networks. It combines ideas from RMSProp (described in lecture) and Momentum.
**How does Adam work? **
- It calculates an exponentially weighted average of past gradients, and stores it in variables $v$ (before bias correction) and $v^{corrected}$ (with bias correction).
- It calculates an exponentially weighted average of the squares of the past gradients, and stores it in variables $s$ (before bias correction) and $s^{corrected}$ (with bias correction).
- It updates parameters in a direction based on combining information from “1” and “2”.
The update rule is, for l=1,...,L : $$\begin{cases} v_{dW^{[l]}} = \beta_1 v_{dW^{[l]}} + (1 - \beta_1) \frac{\partial \mathcal{J} }{ \partial W^{[l]} } \ v^{corrected} {dW^{[l]}} = \frac{v {dW^{[l]}}}{1 - (\beta_1)^t} \ s_{dW^{[l]}} = \beta_2 s_{dW^{[l]}} + (1 - \beta_2) (\frac{\partial \mathcal{J} }{\partial W^{[l]} })^2 \ s^{corrected} {dW^{[l]}} = \frac{s {dW^{[l]}}}{1 - (\beta_2)^t} \ W^{[l]} = W^{[l]} - \alpha \frac{v^{corrected} {dW^{[l]}}}{\sqrt{s^{corrected} {dW^{[l]}}} + \varepsilon} \end{cases}$$
- $t$ counts the number of steps taken of Adam
- $L$ is the number of layers
- $β_1$ and $β_2$ are hyperparameters that control the two exponentially weighted averages.
- $α$ is the learning rate
- $ε$ is a very small number to avoid dividing by zero As usual, we will store all parameters in the parameters dictionary
Exercise : Initialize the Adam variables $v,s$ which keep track of the past information.
Instruction : The variables $v,s$ are python dictionaries that need to be initialized with arrays of zeros. Their keys are the same as for grads, that is: for l=1,...,L :
| 2 3 4 | + str(l+1)] = ... #(numpy array of zeros with the same shape as parameters["W" + str(l+1)]) v["db" + str(l+1)] = ... #(numpy array of zeros with the same shape as parameters["b" + str(l+1)]) s["dW" + str(l+1)] = ... #(numpy array of zeros with the same shape as parameters["W" + str(l+1)]) s["db" + str(l+1)] = ... #(numpy array of zeros with the same shape as parameters["b" + str(l+1)]) |
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 | def initialize_adam(parameters) : """ Initializes v and s as two python dictionaries with: - keys: "dW1", "db1", ..., "dWL", "dbL" - values: numpy arrays of zeros of the same shape as the corresponding gradients/parameters. Arguments: parameters -- python dictionary containing your parameters. parameters["W" + str(l)] = Wl parameters["b" + str(l)] = bl Returns: v -- python dictionary that will contain the exponentially weighted average of the gradient. v["dW" + str(l)] = ... v["db" + str(l)] = ... s -- python dictionary that will contain the exponentially weighted average of the squared gradient. s["dW" + str(l)] = ... s["db" + str(l)] = ... """ L = len(parameters) // 2 # number of layers in the neural networks v = {} s = {} # Initialize v, s. Input: "parameters". Outputs: "v, s". for l in range(L): ### START CODE HERE ### (approx. 4 lines) v['dW' + str(l + 1)] = np.zeros(parameters["W" + str(l + 1)].shape); v['db' + str(l + 1)] = np.zeros(parameters["b" + str(l + 1)].shape); s['dW' + str(l + 1)] = np.zeros(parameters["W" + str(l + 1)].shape); s['db' + str(l + 1)] = np.zeros(parameters["b" + str(l + 1)].shape); ### END CODE HERE ### return v, s |
| 2 3 4 5 6 7 8 9 10 11 | v, s = initialize_adam(parameters) print("v[\"dW1\"] = " + str(v["dW1"])) print("v[\"db1\"] = " + str(v["db1"])) print("v[\"dW2\"] = " + str(v["dW2"])) print("v[\"db2\"] = " + str(v["db2"])) print("s[\"dW1\"] = " + str(s["dW1"])) print("s[\"db1\"] = " + str(s["db1"])) print("s[\"dW2\"] = " + str(s["dW2"])) print("s[\"db2\"] = " + str(s["db2"])) |
| variable | value |
|---|---|
| [[ 0. 0. 0.] [ 0. 0. 0.]] | |
| [[ 0.] [ 0.]] | |
| [[ 0. 0. 0.] [ 0. 0. 0.] [ 0. 0. 0.]] | |
| [[ 0.] [ 0.] [ 0.]] | |
| [[ 0. 0. 0.] [ 0. 0. 0.]] | |
| [[ 0.] [ 0.]] | |
| [[ 0. 0. 0.] [ 0. 0. 0.] [ 0. 0. 0.]] | |
| [[ 0.] [ 0.] [ 0.]] |
Exercise : Now, implement the parameters update with Adam. Recall the general update rule is, for l=1,...,L : $$\begin{cases} v_{dW^{[l]}} = \beta_1 v_{dW^{[l]}} + (1 - \beta_1) \frac{\partial \mathcal{J} }{ \partial W^{[l]} } \ v^{corrected} {dW^{[l]}} = \frac{v {dW^{[l]}}}{1 - (\beta_1)^t} \ s_{dW^{[l]}} = \beta_2 s_{dW^{[l]}} + (1 - \beta_2) (\frac{\partial \mathcal{J} }{\partial W^{[l]} })^2 \ s^{corrected} {dW^{[l]}} = \frac{s {dW^{[l]}}}{1 - (\beta_2)^t} \ W^{[l]} = W^{[l]} - \alpha \frac{v^{corrected} {dW^{[l]}}}{\sqrt{s^{corrected} {dW^{[l]}}} + \varepsilon} \end{cases}$$
Note that the iterator l starts at 0 in the for loop while the first parameters are $W^{[1]}$ and $b^{[1]}$. You need to shift l to l+1 when coding.
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 | def update_parameters_with_adam(parameters, grads, v, s, t, learning_rate = 0.01, beta1 = 0.9, beta2 = 0.999, epsilon = 1e-8): """ Update parameters using Adam Arguments: parameters -- python dictionary containing your parameters: parameters['W' + str(l)] = Wl parameters['b' + str(l)] = bl grads -- python dictionary containing your gradients for each parameters: grads['dW' + str(l)] = dWl grads['db' + str(l)] = dbl v -- Adam variable, moving average of the first gradient, python dictionary s -- Adam variable, moving average of the squared gradient, python dictionary learning_rate -- the learning rate, scalar. beta1 -- Exponential decay hyperparameter for the first moment estimates beta2 -- Exponential decay hyperparameter for the second moment estimates epsilon -- hyperparameter preventing division by zero in Adam updates Returns: parameters -- python dictionary containing your updated parameters v -- Adam variable, moving average of the first gradient, python dictionary s -- Adam variable, moving average of the squared gradient, python dictionary """ L = len(parameters) // 2 # number of layers in the neural networks v_corrected = {} # Initializing first moment estimate, python dictionary s_corrected = {} # Initializing second moment estimate, python dictionary # Perform Adam update on all parameters for l in range(L): # Moving average of the gradients. Inputs: "v, grads, beta1". Output: "v". ### START CODE HERE ### (approx. 2 lines) v['dW' + str(l + 1)] = beta1 * v['dW' + str(l + 1)] + (1 - beta1) * grads['dW' + str(l + 1)]; v['db' + str(l + 1)] = beta1 * v['db' + str(l + 1)] + (1 - beta1) * grads['db' + str(l + 1)]; ### END CODE HERE ### # Compute bias-corrected first moment estimate. Inputs: "v, beta1, t". Output: "v_corrected". ### START CODE HERE ### (approx. 2 lines) v_corrected['dW' + str(l + 1)] = v['dW' + str(l + 1)] / (1 - np.power(beta1, t)); v_corrected['db' + str(l + 1)] = v['db' + str(l + 1)] / (1 - np.power(beta1, t)); ### END CODE HERE ### # Moving average of the squared gradients. Inputs: "s, grads, beta2". Output: "s". ### START CODE HERE ### (approx. 2 lines) s['dW' + str(l + 1)] = beta2 * s['dW' + str(l + 1)] + (1 - beta2) * np.power(grads['dW' + str(l + 1)], 2); s['db' + str(l + 1)] = beta2 * s['db' + str(l + 1)] + (1 - beta2) * np.power(grads['db' + str(l + 1)], 2); ### END CODE HERE ### # Compute bias-corrected second raw moment estimate. Inputs: "s, beta2, t". Output: "s_corrected". ### START CODE HERE ### (approx. 2 lines) s_corrected['dW' + str(l + 1)] = s['dW' + str(l + 1)] / (1 - np.power(beta2, t)); s_corrected['db' + str(l + 1)] = s['db' + str(l + 1)] / (1 - np.power(beta2, t)); ### END CODE HERE ### # Update parameters. Inputs: "parameters, learning_rate, v_corrected, s_corrected, epsilon". Output: "parameters". ### START CODE HERE ### (approx. 2 lines) parameters['W' + str(l + 1)] -= learning_rate * v_corrected['dW' + str(l + 1)] / (np.sqrt(s_corrected['dW' + str(l + 1)]) + epsilon); parameters['b' + str(l + 1)] -= learning_rate * v_corrected['db' + str(l + 1)] / (np.sqrt(s_corrected['db' + str(l + 1)]) + epsilon); ### END CODE HERE ### return parameters, v, s |
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 | parameters, v, s = update_parameters_with_adam(parameters, grads, v, s, t = 2) print("W1 = " + str(parameters["W1"])) print("b1 = " + str(parameters["b1"])) print("W2 = " + str(parameters["W2"])) print("b2 = " + str(parameters["b2"])) print("v[\"dW1\"] = " + str(v["dW1"])) print("v[\"db1\"] = " + str(v["db1"])) print("v[\"dW2\"] = " + str(v["dW2"])) print("v[\"db2\"] = " + str(v["db2"])) print("s[\"dW1\"] = " + str(s["dW1"])) print("s[\"db1\"] = " + str(s["db1"])) print("s[\"dW2\"] = " + str(s["dW2"])) print("s[\"db2\"] = " + str(s["db2"])) |
| variable | value |
|---|---|
| [[ 1.63178673 -0.61919778 -0.53561312] [-1.08040999 0.85796626 -2.29409733]] | |
| [[ 1.75225313] [-0.75376553]] | |
| [[ 0.32648046 -0.25681174 1.46954931] [-2.05269934 -0.31497584 -0.37661299] [ 1.14121081 -1.09245036 -0.16498684]] | |
| [[-0.88529978] [ 0.03477238] [ 0.57537385]] | |
| [[-0.11006192 0.11447237 0.09015907] [ 0.05024943 0.09008559 -0.06837279]] | |
| [[-0.01228902] [-0.09357694]] | |
| [[-0.02678881 0.05303555 -0.06916608] [-0.03967535 -0.06871727 -0.08452056] [-0.06712461 -0.00126646 -0.11173103]] | |
| [[ 0.02344157] [ 0.16598022] [ 0.07420442]] | |
| [[ 0.00121136 0.00131039 0.00081287] [ 0.0002525 0.00081154 0.00046748]] | |
| [[ 1.51020075e-05] [ 8.75664434e-04]] | |
| [[ 7.17640232e-05 2.81276921e-04 4.78394595e-04] [ 1.57413361e-04 4.72206320e-04 7.14372576e-04] [ 4.50571368e-04 1.60392066e-07 1.24838242e-03]] | |
| [[ 5.49507194e-05] [ 2.75494327e-03] [ 5.50629536e-04]] |
You now have three working optimization algorithms (mini-batch gradient descent, Momentum, Adam). Let’s implement a model with each of these optimizers and observe the difference.
5 - Model with different optimization algorithms
Lets use the following “moons” dataset to test the different optimization methods. (The dataset is named “moons” because the data from each of the two classes looks a bit like a crescent-shaped moon.)
| | |
We have already implemented a 3-layer neural network. You will train it with:
- Mini-batch Gradient Descent: it will call your function: update_parameters_with_gd()
- Mini-batch Momentum: it will call your functions: initialize_velocity() and update_parameters_with_momentum()
- Mini-batch Adam: it will call your functions: initialize_adam() and update_parameters_with_adam()
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 | model(X, Y, layers_dims, optimizer, learning_rate = 0.0007, mini_batch_size = 64, beta = 0.9, beta1 = 0.9, beta2 = 0.999, epsilon = 1e-8, num_epochs = 10000, print_cost = True): """ 3-layer neural network model which can be run in different optimizer modes. Arguments: X -- input data, of shape (2, number of examples) Y -- true "label" vector (1 for blue dot / 0 for red dot), of shape (1, number of examples) layers_dims -- python list, containing the size of each layer learning_rate -- the learning rate, scalar. mini_batch_size -- the size of a mini batch beta -- Momentum hyperparameter beta1 -- Exponential decay hyperparameter for the past gradients estimates beta2 -- Exponential decay hyperparameter for the past squared gradients estimates epsilon -- hyperparameter preventing division by zero in Adam updates num_epochs -- number of epochs print_cost -- True to print the cost every 1000 epochs Returns: parameters -- python dictionary containing your updated parameters """ L = len(layers_dims) # number of layers in the neural networks costs = [] # to keep track of the cost t = 0 # initializing the counter required for Adam update seed = 10 # For grading purposes, so that your "random" minibatches are the same as ours # Initialize parameters parameters = initialize_parameters(layers_dims) # Initialize the optimizer if optimizer == "gd": pass # no initialization required for gradient descent elif optimizer == "momentum": v = initialize_velocity(parameters) elif optimizer == "adam": v, s = initialize_adam(parameters) # Optimization loop for i in range(num_epochs): # Define the random minibatches. We increment the seed to reshuffle differently the dataset after each epoch seed = seed + 1 minibatches = random_mini_batches(X, Y, mini_batch_size, seed) for minibatch in minibatches: # Select a minibatch (minibatch_X, minibatch_Y) = minibatch # Forward propagation a3, caches = forward_propagation(minibatch_X, parameters) # Compute cost cost = compute_cost(a3, minibatch_Y) # Backward propagation grads = backward_propagation(minibatch_X, minibatch_Y, caches) # Update parameters if optimizer == "gd": parameters = update_parameters_with_gd(parameters, grads, learning_rate) elif optimizer == "momentum": parameters, v = update_parameters_with_momentum(parameters, grads, v, beta, learning_rate) elif optimizer == "adam": t = t + 1 # Adam counter parameters, v, s = update_parameters_with_adam(parameters, grads, v, s, t, learning_rate, beta1, beta2, epsilon) # Print the cost every 1000 epoch if print_cost and i % 1000 == 0: print ("Cost after epoch %i: %f" %(i, cost)) if print_cost and i % 100 == 0: costs.append(cost) # plot the cost plt.plot(costs) plt.ylabel('cost') plt.xlabel('epochs (per 100)') plt.title("Learning rate = " + str(learning_rate)) plt.show() return parameters |
You will now run this 3 layer neural network with each of the 3 optimization methods.
5.1 Mini-batch Gradient descent
Run the following code to see how the model does with mini-batch gradient descent.
| 2 3 4 5 6 7 8 9 10 11 12 13 | layers_dims = [train_X.shape[0], 5, 2, 1] parameters = model(train_X, train_Y, layers_dims, optimizer = "gd") # Predict predictions = predict(train_X, train_Y, parameters) # Plot decision boundary plt.title("Model with Gradient Descent optimization") axes = plt.gca() axes.set_xlim([-1.5,2.5]) axes.set_ylim([-1,1.5]) plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y) |
5.2 Mini-batch gradient descent with momentum
Run the following code to see how the model does with momentum. Because this example is relatively simple, the gains from using momemtum are small; but for more complex problems you might see bigger gains.
| 2 3 4 5 6 7 8 9 10 11 12 13 | layers_dims = [train_X.shape[0], 5, 2, 1] parameters = model(train_X, train_Y, layers_dims, beta = 0.9, optimizer = "momentum") # Predict predictions = predict(train_X, train_Y, parameters) # Plot decision boundary plt.title("Model with Momentum optimization") axes = plt.gca() axes.set_xlim([-1.5,2.5]) axes.set_ylim([-1,1.5]) plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y) |
5.3 Mini-batch with Adam mode
Run the following code to see how the model does with Adam.
| 2 3 4 5 6 7 8 9 10 11 12 13 | layers_dims = [train_X.shape[0], 5, 2, 1] parameters = model(train_X, train_Y, layers_dims, optimizer = "adam") # Predict predictions = predict(train_X, train_Y, parameters) # Plot decision boundary plt.title("Model with Adam optimization") axes = plt.gca() axes.set_xlim([-1.5,2.5]) axes.set_ylim([-1,1.5]) plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y) |
5.4 Summary
| Gradient descent | 79.7% | oscillations |
| Momentum | 79.7% | oscillations |
| Adam | 94% | smoother |
Momentum usually helps, but given the small learning rate and the simplistic dataset, its impact is almost negligeable. Also, the huge oscillations you see in the cost come from the fact that some minibatches are more difficult thans others for the optimization algorithm.
Adam on the other hand, clearly outperforms mini-batch gradient descent and Momentum. If you run the model for more epochs on this simple dataset, all three methods will lead to very good results. However, you’ve seen that Adam converges a lot faster.
Some advantages of Adam include: - Relatively low memory requirements (though higher than gradient descent and gradient descent with momentum) - Usually works well even with little tuning of hyperparameters (except $α$)
References :
- Adam paper: https://arxiv.org/pdf/1412.6980.pdf

- Post author: SnailDove
- Post link: https://snaildove.github.io/2018/03/02/OptimizationMethods/
- Copyright Notice: All articles in this blog are licensed under CC BY-NC-SA 3.0 unless stating additionally.
Subscribe to the PwC Newsletter
Join the community, edit social preview.

Add a new code entry for this paper
Remove a code repository from this paper.

Mark the official implementation from paper authors
Add a new evaluation result row.
| TASK | DATASET | MODEL | METRIC NAME | METRIC VALUE | GLOBAL RANK | REMOVE |
|---|
Remove a task
Add a method, remove a method, edit datasets, combining optimization methods using an adaptive meta optimizer.
Algorithsm MDPI 2021 · Nicola Landro , Ignazio Gallo , Riccardo La Grassa · Edit social preview
Optimization methods are of great importance for the efficient training of neural networks. There are many articles in the literature that propose particular variants of existing optimizers. In our article, we propose the use of the combination of two very different optimizers that, when used simultaneously, can exceed the performance of the single optimizers in very different problems. We propose a new optimizer called ATMO (AdapTive Meta Optimizers), which integrates two different optimizers simultaneously weighing the contributions of both. Rather than trying to improve each single one, we leverage both at the same time, as a meta-optimizer, by taking the best of both. We have conducted several experiments on the classification of images and text documents, using various types of deep neural models, and we have demonstrated through experiments that the proposed ATMO produces better performance than the single optimizers.
Code Edit Add Remove Mark official
Tasks edit add remove, datasets edit.

Results from the Paper Edit Add Remove
Methods edit add remove.
Gurobi Modeling Examples
Explore our modeling examples for the Gurobi Python API

Target audience
Data scientists, engineers, computer scientists, economists, and in general, professionals with a background in mathematical modeling and a basic knowledge of Python.
Goals of modeling examples
- Illustrate the broad applicability of mathematical optimization.
- Show how to build mathematical optimization models.
These modeling examples are coded using the Gurobi Python API and distributed as Jupyter Notebooks.
These modeling examples illustrate important capabilities of the Gurobi Python API, including adding decision variables, building linear expressions, adding constraints, and adding an objective function. They touch on more advanced features such as generalized constraints, piecewise-linear functions, and multi-objective hierarchical optimization. They also illustrate common constraint types such as “allocation constraints”, “balance constraints”, “sequencing constraints”, “precedence constraints”, and others.
The examples are from different business purposes and reflect different levels of building mathematical optimization models.
Introductory examples
The introductory examples walk you through the process of building a mathematical optimization model. The basic requirements are that you know Python and have a background in a discipline that uses quantitative methods.
- GDD 2023: Intro to Gurobipy: This tutorial was given at the Gurobi Days Digital 2023. It is an introduction to the Gurobi Python API Gurobipy. It walks you through the basics of Gurobipy and explains its usage with some small examples.
- Intro to Mathematical Optimization Modeling: This tutorial discusses the basics of mathematical modeling on the example of a simple assignment problem.
- Optimization 101: This tutorial is based on the Webinar on Optimization 101 for Data Scientists and consists of two modeling sessions with exercises and questions as well as a discussion of a use case.
- Airline Planning After Flight Disruption
- Music Recommendation
- Text Dissimilarity
- Power Generation
Beginner Examples
The notebooks at the beginner level assume you know Python and have some knowledge about building mathematical optimization models.
- 3D Tic-Tac-Toe: This example will show you how a binary programming model can be used to capture simple logical constraints.
- Cell Tower: In this example, you will learn how to define and solve a covering-type problem, namely, how to configure a network of cell towers to provide signal coverage to the largest number of people.
- Curve Fitting: Try this Jupyter Notebook Modeling Example to learn how you can fit a function to a set of observations.
- Facility Location: In this example, we will show you how to tackle a facility location problem that involves determining the number and location of warehouses that are needed to supply a group of supermarkets.
- Fantasy Basketball: This example combines machine learning and optimization modeling in fantasy basketball.
- Food Program: Transporting food in a global transportation network is a challenging undertaking. In this notebook, we will build an optimization model to set up a food supply chain based on real data from the UN World Food Program.
- Market Sharing: In this example, we will show you how to solve a goal programming problem that involves allocating the retailers to two divisions of a company in order to optimize the trade-offs of several market-sharing goals.
- Marketing Campaign Optimization: Companies across almost every industry are looking to optimize their marketing campaigns. In this Jupyter Notebook, we will explore a marketing campaign optimization problem that is common in the banking and financial services industry, which involves determining which products to offer to individual customers in order to maximize total expected profit while satisfying various business constraints.
- Offshore Wind Farming: In this example, we will learn how to solve an offshore wind power generation problem. The goal of the problem is to figure out which underwater cables should be laid to connect an offshore wind farm power network at a minimum cost.
- Supply Network Design 1: Try this Jupyter Notebook Modeling Example to learn how to solve a classic supply network design problem that involves finding the minimum cost flow through a network. We will show you how – given a set of factories, depots, and customers – you can use mathematical optimization to determine the best way to satisfy customer demand while minimizing shipping costs. In part 2, we additionally determine which depots to open or close in order to minimize overall costs.
Intermediate Examples
Examples at the intermediate level assume that you know Python and are familiar with the Gurobi Python API. In addition, you should have knowledge about building mathematical optimization models.
- Agricultural Pricing: Try this example to learn how to use mathematical optimization to tackle a common, but critical agricultural pricing problem: Determining the prices and demand for a country’s dairy products in order to maximize total revenue derived from the sales of those products. You will learn how to model this problem as a quadratic optimization problem using the Gurobi Python API and solve it using the Gurobi Optimizer.
- Linear Regression: In this example, you will learn how to perform linear regression with feature selection using mathematical programming. We will show you how to construct a mixed-integer quadratic programming (MIQP) model of this linear regression problem.
- Car Rental: This notebook will teach you how you can use mathematical optimization to figure out how many cars a car rental company should own and where they should be located every day to maximize weekly profits. Part 2 considers an extension on how mathematical optimization can be used to figure out in which locations a car rental company should expand repair capacity.
- Customer Assignment: This notebook is an intermediate version of the facility location problem. In addition, we show how machine learning can be used in the pre-processing so as to reduce the computational burden of big datasets.
- Economic Planning: In this example, you will discover how mathematical optimization can be used to address a macroeconomic planning problem that a country may face. The goal is to determine different possible growth patterns for the economy.
- Efficiency Analysis: How can mathematical optimization be used to measure the efficiency of an organization? Find out in this example, where you will learn how to formulate an Efficiency Analysis model as a linear programming problem.
- Electrical Power Generation: This model is an example of an electrical power generation problem (also known as a unit commitment problem). It selects an optimal set of power stations to turn on in order to satisfy anticipated power demand over a 24-hour time horizon. In part 2, the model is extended and adds the option of using hydroelectric power plants to satisfy demand.
- Factory Planning: In this example, we create an optimal production plan that maximizes profits. In part 2, we create an optimal production plan that will not only maximize profits but also determine the months in which to perform maintenance operations on the machines.
- Food Manufacturing: You will learn how to create an optimal multi-period production plan for a product that requires a number of ingredients – each of which has different costs, restrictions, and features. In part 2, additional constraints are considered that change the problem type from a linear program (LP) problem to a mixed-integer program (MIP) problem, making it harder to solve.
- Logical Design: In this example, you will learn how to solve a logical design problem, which involves constructing a circuit using the minimum number of NOR gates (devices with two inputs and one output) that will perform the logical function specified by a truth table.
- Mining: In this example, you will learn how to model and solve a multi-period production planning problem that involves optimizing the operations of a group of mines over a five-year period.
- Opencast Mining: This notebook shows a mathematical optimization problem to identify which excavation locations to choose in order to maximize the gross margins of extracting ore.
- Power Generation: Assume that we know the set of all available power plants and the demand for power for each hour of a day. We want to create a schedule to decide how much power each plant should generate, and when to switch the plants “on” and “off” in order to minimize the overall costs.
- Refinery: This model is an example of a production planning problem where decisions must be made regarding which products to produce and which resources to use.
- Technician Routing and Scheduling: Try this modeling example to discover how mathematical optimization can help telecommunications firms automate and improve their technician assignment, scheduling, and routing decisions in order to ensure the highest levels of customer satisfaction. You will learn how to formulate a multi-depot vehicle routing problem with time windows constraints.
Advanced Examples
For modeling examples at the advanced level, we assume that you know Python and the Gurobi Python API and that you have advanced knowledge of building mathematical optimization models. Typically, the objective function and/or constraints of these examples are complex or require advanced features of the Gurobi Python API.
- Constraint Optimization: In this example, we consider a constraint of an integer programming model where all the decision variables in the constraint are binary, the goal is to find another constraint involving the same binary variables that is logically equivalent to the original constraint, but that has the smallest possible absolute value of the right-hand side.
- Decentralization Planning: This model is an advanced version of a facility location problem. Given a set of departments of a company and potential cities where these departments can be located, we want to determine the “best” location of each department in order to maximize gross margins.
- Farm Planning: This is an example of an advanced production planning problem.
- Lost Luggage Distribution: This is an example of a vehicle routing problem with time windows. It involves helping a company figure out the minimum number of vans required to deliver pieces of lost or delayed baggage to their rightful owners and determining the optimal assignment of vans to customers.
- Manpower Planning: This notebook solves a staffing planning problem where choices must be made regarding recruitment, training, redundancy, and scheduling of staff.
- Milk Collection: This is an example of a capacitated vehicle routing problem. With only one tanker truck with limited capacity, you will need to determine the best possible route for the tanker to take to collect milk every day from a set of farms.
- Portfolio Selection Optimization: This model is an example of the classic Markowitz portfolio selection optimization model. We want to find the fraction of the portfolio to invest among a set of stocks that balances risk and return. It is a Quadratic Programming (QP) model with vector and matrix data for returns and risk, respectively.
- Pooling: Companies across numerous industries – including petrochemical refining, wastewater treatment, and mining – use mathematical optimization to solve the pooling problem. This problem can be regarded as a generalization of the minimum-cost flow problem and the blending problem.
- Protein Comparison: You will learn how to model the protein comparison problem as a quadratic assignment problem. It involves measuring the similarities of two proteins.
- Protein Folding: The problem pertains to a protein, which consists of a chain of amino acids. The objective is to predict the optimum folding of the chain.
- Traveling Salesman: This notebook covers one of the most famous combinatorial optimization problems in existence: the Traveling Salesman Problem (TSP). The goal of the TSP – to find the shortest possible route that visits each city once and returns to the original city – is simple, but solving the problem is a complex and challenging endeavor. This example uses the callback feature of Gurobi.
- Workforce Scheduling: In this notebook, we demonstrate how you can use mathematical optimization to generate an optimal workforce schedule that minimizes the number of temporary workers your company needs to hire and maximizes employee fairness. The problem is formulated as a multi-objective mixed-integer-programming (MIP) model and uses the multiple objectives feature of Gurobi.
- Yield Management: In this example, we will show you how an airline can use AI technology to devise an optimal seat pricing strategy. You will learn how to formulate this Yield Management Problem as a three-period stochastic programming problem.
Examples via Business Needs
- Marketing Campaign Optimization (beginner)
- Supply Network Design (beginner)
- Technician Routing and Scheduling (intermediate)
- Manpower Planning (advanced)
- Workforce Scheduling (advanced)
- Covid19 Facility Optimization (beginner)
- Yield Management (advanced)
- Price Optimization (introductory)
- Music Recommendation (introductory)
- Fantasy Basketball (beginner)
- Agricultural Pricing (intermediate)
- Linear Regression (intermediate)
- Food Program (beginner)
- Car Rental (intermediate)
- Economic Planning (intermediate)
- Factory Planning (intermediate)
- Food Manufacturing (intermediate)
- Farm Planning (advanced)
- Cell Tower (beginner)
- Facility Location (beginner)
- Customer Assignment (intermediate)
- Opencast Mining (intermediate)
- Decentralization Planning (advanced)
- Traveling Salesman (advanced)
- Airline Planning After Flight Disruption (introductory)
- Power Generation (intermediate)
- Portfolio Selection Optimization (advanced)
- Efficiency Analysis (intermediate)
- Electrical Power Generation (intermediate)
- Mining (intermediate)
- Refinery (intermediate)
- Curve Fitting (beginner)
- Constraint Optimization (intermediate)
- Lost Luggage Distribution (advanced)
- Milk Collection (advanced)
- Market Sharing (beginner)
It is also possible to browse through the examples w.r.t. difficulty level and business needs on the Gurobi website .
Run on Google Colab
You can access all the examples in Google Colab, which is a free, online Jupyter Notebook environment that allows you to write and execute Python code through your browser. You will need to be signed into a Google account to execute the notebooks. But you do not need an account if you just want to look at the notebooks. For each example, the respective colab link is given in the readme:
- To run the example the first time, choose “Runtime” and then click “Run all”.
- All the cells in the Jupyter Notebook will be executed.
- The example will install the gurobipy package. The Gurobi pip package includes a size-limited trial license equivalent to the Gurobi “online course” license. For most of the notebooks, this restricted license is sufficient to run them. For others, you will need a full license, see the license section below.
- You can also modify and re-run individual cells.
- For subsequent runs, choose “Runtime” and click “Restart and run all”.
- The Gurobi Optimizer will find the optimal solution of the modeling example. Check out the Colab Getting Started Guide for full details on how to use Colab Notebooks as well as create your own.
Run locally
- Clone the repository containing all examples or download it by clicking here
- Start Jupyter Notebook Server
- Open the particular notebook in Jupyter Notebook.
- The notebook will install the gurobipy package and other dependencies. The Gurobi pip package includes a size-limited trial license equivalent to the Gurobi “online course” license. For most of the notebooks, this restricted license is sufficient. For others, you will need a full license.
In order to run the Jupyter Notebooks you will need a Gurobi license. Most of the notebooks can be run using the “online course” license version of Gurobi. This is a limited license and restricts the number of allowed variables and constraints. This restricted license comes also with the gurobipy package when installing it via pip or conda. You can also request a full license, i.e., an evaluation license as a commercial user , or download a free license as an academic user . The latter two license types allow you to run all notebooks. All licenses can also be requested in the Gurobi User Portal after registering for a Gurobi account .
Download the repository
You can download the repository containing all examples by clicking here .
Index of modeling examples
- 3D Tic-Tac-Toe
- Agricultural Pricing
- Burrito Game
- Cutting Stock
- Constraint Optimization
- Covid19 Facility Optimization
- Curve Fitting
- Customer Assignment
- Decentralization Planning
- Drone Network
- Economic Planning
- Efficiency Analysis
- Electrical Power Generation
- Facility Location
- Factory Planning
- Fantasy Basketball
- Farm Planning
- Food Manufacturing
- Food Program
- GDD 2023: Intro to Gurobipy
- Intro to Mathematical Optimization Modeling / MILP Tutorial
- Linear Regression
- Logical Design
- Lost Luggage Distribution
- Manpower Planning
- Market Sharing
- Marketing Campaign Optimization
- Milk Collection
- Offshore Wind Farming
- Opencast Mining
- Optimization 101
- Portfolio Selection Optimization
- Price Optimization
- Protein Comparison
- Protein Folding
- Supply Network Design
- Technician Routing and Scheduling
- Traveling Salesman
- Workforce Scheduling
- Yield Management
These modeling examples are distributed under the Apache 2.0 license © Gurobi Optimization, LLC

IMAGES
VIDEO
COMMENTS
Contribute to kenhding/Coursera development by creating an account on GitHub. My course work solutions and quiz answers. Contribute to kenhding/Coursera development by creating an account on GitHub. ... DL_C2_week2_Optimization_methods.ipynb. Top. File metadata and controls. Preview. Code. Blame. 2449 lines (2449 loc) · 596 KB. Raw. Footer ...
In this notebook, you will learn more advanced optimization methods that can speed up learning and perhaps even get you to a better final value for the cost function. Having a good optimization algorithm can be the difference between waiting days vs. just a few hours to get a good result. Gradient descent goes "downhill" on a cost function J ...
In this notebook, you will learn more advanced optimization methods that can speed up learning and perhaps even get you to a better final value for the cost function. Having a good optimization algorithm can be the difference between waiting days vs. just a few hours to get a good result. ... Updates to Assignment
This repository contains the programming assignments and slides from the deep learning course from coursera offered by deeplearning.ai - gmortuza/Deep-Learning-Specialization Skip to content Navigation Menu
Optimization for Decision Science. #. Computational optimization is essential to many subdomains of engineering, science, and business such as design, control, operations, supply chains, game theory, data science/analytics, and machine learning. This course provides a practical introduction to models, algorithms, and modern software for large ...
Root finding with Newton's method; Adapting Newton's method for optimization; Second derivatives and Hessians; Multivariate Newton's method; Lab: Optimization Using Newton's Method; Quiz: Optimization in Neural Networks and Newton's Method; Programming Assignment: Neural Network with Two Layers
This website offers an open and free introductory course on optimization for machine learning. The course is constructed holistically and as self-contained as possible, in order to cover most optimization principles and methods that are relevant for optimization. This course is recommended as an introductory graduate-level course for Master's ...
To demonstrate the minimization function, consider the problem of minimizing the Rosenbrock function of N variables: f(x) = N − 1 ∑ i = 1100(xi + 1 − x2i)2 + (1 − xi)2. The minimum value of this function is 0 which is achieved when xi = 1. Note that the Rosenbrock function and its derivatives are included in scipy.optimize.
Remember different optimization methods such as (Stochastic) Gradient Descent, Momentum, RMSProp and Adam; Use random mini-batches to accelerate the convergence and improve the optimization; Know the benefits of learning rate decay and apply it to your optimization; Assignment of Week 2. Quiz 2: Optimization algorithms; Programming Assignment ...
In this notebook, you'll gain skills with some more advanced optimization methods that can speed up learning and perhaps even get you to a better final value for the cost function. Having a good optimization algorithm can be the difference between waiting days vs. just a few hours to get a good result. By the end of this notebook, you'll be ...
This course is created by deeplearning.ai. Master Deep Learning, and Break into AI - Qian-Han/coursera-Deep-Learning-Specialization
GitHub Gist: instantly share code, notes, and snippets. GitHub Gist: instantly share code, notes, and snippets. Skip to content. ... wenxin-liu / Optimization Using Gradient Descent: Linear Regression.ipynb. Created April 1, 2023 15:56. Show Gist options. Download ZIP
Notes, programming assignments and quizzes from all courses within the Coursera Deep Learning specialization offered by deeplearning.ai: (i) Neural Networks and Deep Learning; (ii) Improving Deep Neural Networks: Hyperparameter tuning, Regularization and Optimization; (iii) Structuring Machine Learning Projects; (iv) Convolutional Neural Networks; (v) Sequence Models - amanchadha/coursera-deep ...
View on GitHub Neural Networks and Deep Learning. In this course, you will learn the foundations of deep learning. When you finish this class, you will: Understand the major technology trends driving Deep Learning. Be able to build, train and apply fully connected deep neural networks. Know how to implement efficient (vectorized) neural networks.
These are my personal programming assignments at the 2nd week after studying the course Improving Deep Neural Networks: Hyperparameter tuning, Regularization and Optimization and the copyright belongs to deeplearning.ai. Optimization Methods. Until now, you've always used Gradient Descent to update the parameters and minimize the cost.
To associate your repository with the optimization-methods topic, visit your repo's landing page and select "manage topics." GitHub is where people build software. More than 100 million people use GitHub to discover, fork, and contribute to over 420 million projects.
Combining Optimization Methods Using an Adaptive Meta Optimizer. Optimization methods are of great importance for the efficient training of neural networks. There are many articles in the literature that propose particular variants of existing optimizers. In our article, we propose the use of the combination of two very different optimizers ...
The introductory examples walk you through the process of building a mathematical optimization model. The basic requirements are that you know Python and have a background in a discipline that uses quantitative methods. GDD 2023: Intro to Gurobipy: This tutorial was given at the Gurobi Days Digital 2023. It is an introduction to the Gurobi ...
lauraminicucci / Improving-Deep-Neural-Networks-Hyperparameter-tuning-Regularization-and-Optimization Public Notifications You must be signed in to change notification settings Fork 0
You signed in with another tab or window. Reload to refresh your session. You signed out in another tab or window. Reload to refresh your session. You switched accounts on another tab or window.
Programming Assignment for the course "Improving Deep Neural Networks: Hyperparameter tuning, Regularization and Optimization" in Coursera License GPL-3.0 license
Programming assignment covering data normalization techniques and ndarray creation in Python, including experimentation with methods and functions.n of ndarray and experimenting with ithe methods a...
You signed in with another tab or window. Reload to refresh your session. You signed out in another tab or window. Reload to refresh your session. You switched accounts on another tab or window.