- Technical Support
- Find My Rep

You are here
How to get published.
You believe your research will make a contribution to your field, and you’re ready to share it with your peers far and wide, but how do you go about getting it published, and what exactly does that involve?
If this is you, this page is a great place to start. Here you’ll find guidance to taking those first steps towards publication with confidence. From what to consider when choosing a journal, to how to submit an article and what happens next.
Getting started
Choosing the right journal for you.
Submitting your article to a Sage journal
Promoting your article
Related resources you may find useful.

How to Get Your Journal Article Published guide
Our handy guide is a quick overview covering the publishing process from preparing your article and choosing a journal, to publication (5 minute read).
View the How to Get Your Journal Article Published guide

How to Get Published webinars
Free 1 hour monthly How to Get Published webinars cover topics including writing an article, navigating the peer review process, and what exactly it means when you hear “open access.” Join fellow researchers and expert speakers live, or watch our library of recordings on a variety of topics.
Browse our webinars

Sage Perspectives blog
Looking for tips on how to make sure your article goes smoothly through the peer review process, or how to write the right title for your article?
Read our blog

Sage Campus courses
Want something a bit more in-depth? Sage Campus courses are short and interactive (around 2 hours each) and cover a range of skills, including how to get published. Your library may already subscribe to the modules, or you may want to recommend that they do. Meanwhile, you can utilize the free modules.
Explore Sage Campus

Each journal has its own Aims & Scope, so the acceptance of articles is not just about quality, but also about being a good fit. Does your work reflect the scope of the Journal? Is Open Access important to you, and does the Journal have an Open Access model available? What is the readership of the Journal, and is that readership the right audience for your work? Researching the best match for your manuscript will significantly improve your chances of being accepted.
Watch our 2 minute video

If you already know in which Sage journal you’d like to publish your work, search for it and check the manuscript submission guidelines to make sure it is a good match. Or use the Sage Journal Recommender to tell us your article title and subjects and see which journals are a potential home for your manuscript. Be prepared to adjust your manuscript to match the scope and style of the desired journal.
Find journals with the Sage Journal Recommender or browse all Sage journals

Professional presentation of your work includes a precise and clear writing style, avoiding accidental plagiarism, and formatting your article to meet the criteria of your chosen journal. All of these take time and may not be skills inherent to your field of research. Sage Author Services can help you to prepare your manuscript to comply with these and other related standards, which could significantly improve your chance of acceptance.
Visit Sage Author Services
Submitting your article to a Sage journal
You’ve identified the right journal; now you need to make sure your manuscript is the perfect fit. Following the author guidelines can be the difference between possible acceptance and rejection, so it’s definitely worth following the required guidelines. We’ve a selection of resources and guides to help:
Watch How to Get Published: Submitting Your Paper (2 minute video)
Read our Article Submission infographic , a quick reminder of essentials
Here you’ll find chapter and verse on all aspects of our Manuscript Submission Guidelines
Ready to submit? Our online Submission Checklist will help you do a final check before sending your article to us.
Each journal retains editorial independence, which means their Guidelines will vary, so do go to the home page of your chosen journal to check anything you should be aware of. You can submit your article there too.

The academic world is crowded, how can you make your article stand out? If you are active on social media platforms, telling your followers about your article is one of the simplest and most effective things you can do.

Between us, we can improve the chances of your article being found, read, downloaded and cited – of your article and you making an impact. Our tips and guidance will show you how to promote your article alongside building your academic profile.
Read our tips on how to maximize your impact

- Open access at Sage
- Top reasons to publish with Sage
- How to do research and get published webinar series
- How to get published for librarians
- Sage Author Services
- Manuscript submission guidelines
- During peer review
- During and post publication
- Help and support
- Journal Editor Gateway
- Journal Reviewer Gateway
- Ethics & Responsibility
- Sage Editorial Policies
- Publication Ethics Policies
- Sage Chinese Author Gateway 中国作者资源
Unfortunately we don't fully support your browser. If you have the option to, please upgrade to a newer version or use Mozilla Firefox , Microsoft Edge , Google Chrome , or Safari 14 or newer. If you are unable to, and need support, please send us your feedback .
We'd appreciate your feedback. Tell us what you think! opens in new tab/window
7 steps to publishing in a scientific journal
April 5, 2021 | 10 min read
By Aijaz Shaikh, PhD
Before you hit “submit,” here’s a checklist (and pitfalls to avoid)
As scholars, we strive to do high-quality research that will advance science. We come up with what we believe are unique hypotheses, base our work on robust data and use an appropriate research methodology. As we write up our findings, we aim to provide theoretical insight, and share theoretical and practical implications about our work. Then we submit our manuscript for publication in a peer-reviewed journal. For many, this is the hardest part of research. In my seven years of research and teaching, I have observed several shortcomings in the manuscript preparation and submission process that often lead to research being rejected for publication. Being aware of these shortcomings will increase your chances of having your manuscript published and also boost your research profile and career progression.

Dr Aijaz Shaikh gives a presentation.
In this article, intended for doctoral students and other young scholars, I identify common pitfalls and offer helpful solutions to prepare more impactful papers. While there are several types of research articles, such as short communications, review papers and so forth, these guidelines focus on preparing a full article (including a literature review), whether based on qualitative or quantitative methodology, from the perspective of the management, education, information sciences and social sciences disciplines.
Writing for academic journals is a highly competitive activity, and it’s important to understand that there could be several reasons behind a rejection. Furthermore, the journal peer-review process is an essential element of publication because no writer could identify and address all potential issues with a manuscript.
1. Do not rush submitting your article for publication.
In my first article for Elsevier Connect – “Five secrets to surviving (and thriving in) a PhD program” – I emphasized that scholars should start writing during the early stages of your research or doctoral study career. This secret does not entail submitting your manuscript for publication the moment you have crafted its conclusion. Authors sometimes rely on the fact that they will always have an opportunity to address their work’s shortcomings after the feedback received from the journal editor and reviewers has identified them.
A proactive approach and attitude will reduce the chance of rejection and disappointment. In my opinion, a logical flow of activities dominates every research activity and should be followed for preparing a manuscript as well. Such activities include carefully re-reading your manuscript at different times and perhaps at different places. Re-reading is essential in the research field and helps identify the most common problems and shortcomings in the manuscript, which might otherwise be overlooked. Second, I find it very helpful to share my manuscripts with my colleagues and other researchers in my network and to request their feedback. In doing so, I highlight any sections of the manuscript that I would like reviewers to be absolutely clear on.
2. Select an appropriate publication outlet.
I also ask colleagues about the most appropriate journal to submit my manuscript to; finding the right journal for your article can dramatically improve the chances of acceptance and ensure it reaches your target audience.
Elsevier provides an innovative Journal Finder opens in new tab/window search facility on its website. Authors enter the article title, a brief abstract and the field of research to get a list of the most appropriate journals for their article. For a full discussion of how to select an appropriate journal see Knight and Steinbach (2008).
Less experienced scholars sometimes choose to submit their research work to two or more journals at the same time. Research ethics and policies of all scholarly journals suggest that authors should submit a manuscript to only one journal at a time. Doing otherwise can cause embarrassment and lead to copyright problems for the author, the university employer and the journals involved.
3. Read the aims and scope and author guidelines of your target journal carefully.
Once you have read and re-read your manuscript carefully several times, received feedback from your colleagues, and identified a target journal, the next important step is to read the aims and scope of the journals in your target research area. Doing so will improve the chances of having your manuscript accepted for publishing. Another important step is to download and absorb the author guidelines and ensure your manuscript conforms to them. Some publishers report that one paper in five does not follow the style and format requirements of the target journal, which might specify requirements for figures, tables and references.
Rejection can come at different times and in different formats. For instance, if your research objective is not in line with the aims and scope of the target journal, or if your manuscript is not structured and formatted according to the target journal layout, or if your manuscript does not have a reasonable chance of being able to satisfy the target journal’s publishing expectations, the manuscript can receive a desk rejection from the editor without being sent out for peer review. Desk rejections can be disheartening for authors, making them feel they have wasted valuable time and might even cause them to lose enthusiasm for their research topic. Sun and Linton (2014), Hierons (2016) and Craig (2010) offer useful discussions on the subject of “desk rejections.”
4. Make a good first impression with your title and abstract.
The title and abstract are incredibly important components of a manuscript as they are the first elements a journal editor sees. I have been fortunate to receive advice from editors and reviewers on my submissions, and feedback from many colleagues at academic conferences, and this is what I’ve learned:
The title should summarize the main theme of the article and reflect your contribution to the theory.
The abstract should be crafted carefully and encompass the aim and scope of the study; the key problem to be addressed and theory; the method used; the data set; key findings; limitations; and implications for theory and practice.
Dr. Angel Borja goes into detail about these components in “ 11 steps to structuring a science paper editors will take seriously .”
Learn more in Elsevier's free Researcher Academy opens in new tab/window
5. Have a professional editing firm copy-edit (not just proofread) your manuscript, including the main text, list of references, tables and figures.
The key characteristic of scientific writing is clarity. Before submitting a manuscript for publication, it is highly advisable to have a professional editing firm copy-edit your manuscript. An article submitted to a peer-reviewed journal will be scrutinized critically by the editorial board before it is selected for peer review. According to a statistic shared by Elsevier, between 30 percent and 50 percent of articles submitted to Elsevier journals are rejected before they even reach the peer-review stage, and one of the top reasons for rejection is poor language. A properly written, edited and presented text will be error free and understandable and will project a professional image that will help ensure your work is taken seriously in the world of publishing. On occasion, the major revisions conducted at the request of a reviewer will necessitate another round of editing. Authors can facilitate the editing of their manuscripts by taking precautions at their end. These include proofreading their own manuscript for accuracy and wordiness (avoid unnecessary or normative descriptions like “it should be noted here” and “the authors believe) and sending it for editing only when it is complete in all respects and ready for publishing. Professional editing companies charge hefty fees, and it is simply not financially viable to have them conduct multiple rounds of editing on your article. Applications like the spelling and grammar checker in Microsoft Word or Grammarly are certainly worth applying to your article, but the benefits of proper editing are undeniable. For more on the difference between proofreading and editing, see the description in Elsevier’s WebShop.
6. Submit a cover letter with the manuscript.
Never underestimate the importance of a cover letter addressed to the editor or editor-in-chief of the target journal. Last year, I attended a conference in Boston. A “meet the editors” session revealed that many submissions do not include a covering letter, but the editors-in-chief present, who represented renewed and ISI-indexed Elsevier journals, argued that the cover letter gives authors an important opportunity to convince them that their research work is worth reviewing.
Accordingly, the content of the cover letter is also worth spending time on. Some inexperienced scholars paste the article’s abstract into their letter thinking it will be sufficient to make the case for publication; it is a practice best avoided. A good cover letter first outlines the main theme of the paper; second, argues the novelty of the paper; and third, justifies the relevance of the manuscript to the target journal. I would suggest limiting the cover letter to half a page. More importantly, peers and colleagues who read the article and provided feedback before the manuscript’s submission should be acknowledged in the cover letter.
7. Address reviewer comments very carefully.
Editors and editors-in-chief usually couch the acceptance of a manuscript as subject to a “revise and resubmit” based on the recommendations provided by the reviewer or reviewers. These revisions may necessitate either major or minor changes in the manuscript. Inexperienced scholars should understand a few key aspects of the revision process. First, it important to address the revisions diligently; second, is imperative to address all the comments received from the reviewers and avoid oversights; third, the resubmission of the revised manuscript must happen by the deadline provided by the journal; fourth, the revision process might comprise multiple rounds. The revision process requires two major documents. The first is the revised manuscript highlighting all the modifications made following the recommendations received from the reviewers. The second is a letter listing the authors’ responses illustrating they have addressed all the concerns of the reviewers and editors. These two documents should be drafted carefully. The authors of the manuscript can agree or disagree with the comments of the reviewers (typically agreement is encouraged) and are not always obliged to implement their recommendations, but they should in all cases provide a well-argued justification for their course of action.
Given the ever increasing number of manuscripts submitted for publication, the process of preparing a manuscript well enough to have it accepted by a journal can be daunting. High-impact journals accept less than 10 percent of the articles submitted to them, although the acceptance ratio for special issues or special topics sections is normally over 40 percent. Scholars might have to resign themselves to having their articles rejected and then reworking them to submit them to a different journal before the manuscript is accepted.
The advice offered here is not exhaustive but it’s also not difficult to implement. These recommendations require proper attention, planning and careful implementation; however, following this advice could help doctoral students and other scholars improve the likelihood of getting their work published, and that is key to having a productive, exciting and rewarding academic career.
Acknowledgements
I would like to thank Professor Heikki Karjaluoto, Jyväskylä University School of Business and Economics for providing valuable feedback on this article.
Sun, H., & Linton, J. D. (2014).
Structuring papers for success: Making your paper more like a high impact publication than a desk reject opens in new tab/window
Technovation.
Craig, J. B. (2010).
Desk rejection: How to avoid being hit by a returning boomerang opens in new tab/window
Family Business Review
Hierons, R. M. (2016).
The dreaded desk reject opens in new tab/window
, Software Testing, Verification and Reliability .
Borja, A (2014):
11 steps to structuring a science paper editors will take seriously
Elsevier Connect
Knight, L. V., & Steinbach, T. A. (2008).
Selecting an appropriate publication outlet: a comprehensive model of journal selection criteria for researchers in a broad range of academic disciplines opens in new tab/window
, International Journal of Doctoral Studies .
Tewin, K. (2015).
How to Better Proofread An Article in 6 Simple Steps opens in new tab/window ,
Day, R, & Gastel, B: How to write and publish a scientific paper. Cambridge University Press (2012)
Contributor
Aijaz shaikh, phd.
- PRO Courses Guides New Tech Help Pro Expert Videos About wikiHow Pro Upgrade Sign In
- EDIT Edit this Article
- EXPLORE Tech Help Pro About Us Random Article Quizzes Request a New Article Community Dashboard This Or That Game Happiness Hub Popular Categories Arts and Entertainment Artwork Books Movies Computers and Electronics Computers Phone Skills Technology Hacks Health Men's Health Mental Health Women's Health Relationships Dating Love Relationship Issues Hobbies and Crafts Crafts Drawing Games Education & Communication Communication Skills Personal Development Studying Personal Care and Style Fashion Hair Care Personal Hygiene Youth Personal Care School Stuff Dating All Categories Arts and Entertainment Finance and Business Home and Garden Relationship Quizzes Cars & Other Vehicles Food and Entertaining Personal Care and Style Sports and Fitness Computers and Electronics Health Pets and Animals Travel Education & Communication Hobbies and Crafts Philosophy and Religion Work World Family Life Holidays and Traditions Relationships Youth
- Browse Articles
- Learn Something New
- Quizzes Hot
- Happiness Hub
- This Or That Game
- Train Your Brain
- Explore More
- Support wikiHow
- About wikiHow
- Log in / Sign up
- Education and Communications
- College University and Postgraduate
- Academic Writing
- Research Papers
How to Write and Publish Your Research in a Journal
Last Updated: May 26, 2024 Fact Checked
Choosing a Journal
Writing the research paper, editing & revising your paper, submitting your paper, navigating the peer review process, research paper help.
This article was co-authored by Matthew Snipp, PhD and by wikiHow staff writer, Cheyenne Main . C. Matthew Snipp is the Burnet C. and Mildred Finley Wohlford Professor of Humanities and Sciences in the Department of Sociology at Stanford University. He is also the Director for the Institute for Research in the Social Science’s Secure Data Center. He has been a Research Fellow at the U.S. Bureau of the Census and a Fellow at the Center for Advanced Study in the Behavioral Sciences. He has published 3 books and over 70 articles and book chapters on demography, economic development, poverty and unemployment. He is also currently serving on the National Institute of Child Health and Development’s Population Science Subcommittee. He holds a Ph.D. in Sociology from the University of Wisconsin—Madison. There are 13 references cited in this article, which can be found at the bottom of the page. This article has been fact-checked, ensuring the accuracy of any cited facts and confirming the authority of its sources. This article has been viewed 705,728 times.
Publishing a research paper in a peer-reviewed journal allows you to network with other scholars, get your name and work into circulation, and further refine your ideas and research. Before submitting your paper, make sure it reflects all the work you’ve done and have several people read over it and make comments. Keep reading to learn how you can choose a journal, prepare your work for publication, submit it, and revise it after you get a response back.
Things You Should Know
- Create a list of journals you’d like to publish your work in and choose one that best aligns with your topic and your desired audience.
- Prepare your manuscript using the journal’s requirements and ask at least 2 professors or supervisors to review your paper.
- Write a cover letter that “sells” your manuscript, says how your research adds to your field and explains why you chose the specific journal you’re submitting to.

- Ask your professors or supervisors for well-respected journals that they’ve had good experiences publishing with and that they read regularly.
- Many journals also only accept specific formats, so by choosing a journal before you start, you can write your article to their specifications and increase your chances of being accepted.
- If you’ve already written a paper you’d like to publish, consider whether your research directly relates to a hot topic or area of research in the journals you’re looking into.

- Review the journal’s peer review policies and submission process to see if you’re comfortable creating or adjusting your work according to their standards.
- Open-access journals can increase your readership because anyone can access them.

- Scientific research papers: Instead of a “thesis,” you might write a “research objective” instead. This is where you state the purpose of your research.
- “This paper explores how George Washington’s experiences as a young officer may have shaped his views during difficult circumstances as a commanding officer.”
- “This paper contends that George Washington’s experiences as a young officer on the 1750s Pennsylvania frontier directly impacted his relationship with his Continental Army troops during the harsh winter at Valley Forge.”

- Scientific research papers: Include a “materials and methods” section with the step-by-step process you followed and the materials you used. [5] X Research source
- Read other research papers in your field to see how they’re written. Their format, writing style, subject matter, and vocabulary can help guide your own paper. [6] X Research source

- If you’re writing about George Washington’s experiences as a young officer, you might emphasize how this research changes our perspective of the first president of the U.S.
- Link this section to your thesis or research objective.
- If you’re writing a paper about ADHD, you might discuss other applications for your research.

- Scientific research papers: You might include your research and/or analytical methods, your main findings or results, and the significance or implications of your research.
- Try to get as many people as you can to read over your abstract and provide feedback before you submit your paper to a journal.

- They might also provide templates to help you structure your manuscript according to their specific guidelines. [11] X Research source

- Not all journal reviewers will be experts on your specific topic, so a non-expert “outsider’s perspective” can be valuable.

- If you have a paper on the purification of wastewater with fungi, you might use both the words “fungi” and “mushrooms.”
- Use software like iThenticate, Turnitin, or PlagScan to check for similarities between the submitted article and published material available online. [15] X Research source

- Header: Address the editor who will be reviewing your manuscript by their name, include the date of submission, and the journal you are submitting to.
- First paragraph: Include the title of your manuscript, the type of paper it is (like review, research, or case study), and the research question you wanted to answer and why.
- Second paragraph: Explain what was done in your research, your main findings, and why they are significant to your field.
- Third paragraph: Explain why the journal’s readers would be interested in your work and why your results are important to your field.
- Conclusion: State the author(s) and any journal requirements that your work complies with (like ethical standards”).
- “We confirm that this manuscript has not been published elsewhere and is not under consideration by another journal.”
- “All authors have approved the manuscript and agree with its submission to [insert the name of the target journal].”

- Submit your article to only one journal at a time.
- When submitting online, use your university email account. This connects you with a scholarly institution, which can add credibility to your work.

- Accept: Only minor adjustments are needed, based on the provided feedback by the reviewers. A first submission will rarely be accepted without any changes needed.
- Revise and Resubmit: Changes are needed before publication can be considered, but the journal is still very interested in your work.
- Reject and Resubmit: Extensive revisions are needed. Your work may not be acceptable for this journal, but they might also accept it if significant changes are made.
- Reject: The paper isn’t and won’t be suitable for this publication, but that doesn’t mean it might not work for another journal.

- Try organizing the reviewer comments by how easy it is to address them. That way, you can break your revisions down into more manageable parts.
- If you disagree with a comment made by a reviewer, try to provide an evidence-based explanation when you resubmit your paper.

- If you’re resubmitting your paper to the same journal, include a point-by-point response paper that talks about how you addressed all of the reviewers’ comments in your revision. [22] X Research source
- If you’re not sure which journal to submit to next, you might be able to ask the journal editor which publications they recommend.

Expert Q&A
You might also like.

- If reviewers suspect that your submitted manuscript plagiarizes another work, they may refer to a Committee on Publication Ethics (COPE) flowchart to see how to move forward. [23] X Research source Thanks Helpful 0 Not Helpful 0

- ↑ https://www.wiley.com/en-us/network/publishing/research-publishing/choosing-a-journal/6-steps-to-choosing-the-right-journal-for-your-research-infographic
- ↑ https://link.springer.com/article/10.1007/s13187-020-01751-z
- ↑ https://libguides.unomaha.edu/c.php?g=100510&p=651627
- ↑ https://www.canberra.edu.au/library/start-your-research/research_help/publishing-research
- ↑ https://writingcenter.fas.harvard.edu/conclusions
- ↑ https://writing.wisc.edu/handbook/assignments/writing-an-abstract-for-your-research-paper/
- ↑ https://www.springer.com/gp/authors-editors/book-authors-editors/your-publication-journey/manuscript-preparation
- ↑ https://apus.libanswers.com/writing/faq/2391
- ↑ https://academicguides.waldenu.edu/library/keyword/search-strategy
- ↑ https://ifis.libguides.com/journal-publishing-guide/submitting-your-paper
- ↑ https://www.springer.com/kr/authors-editors/authorandreviewertutorials/submitting-to-a-journal-and-peer-review/cover-letters/10285574
- ↑ https://www.apa.org/monitor/sep02/publish.aspx
- ↑ Matthew Snipp, PhD. Research Fellow, U.S. Bureau of the Census. Expert Interview. 26 March 2020.
About This Article

To publish a research paper, ask a colleague or professor to review your paper and give you feedback. Once you've revised your work, familiarize yourself with different academic journals so that you can choose the publication that best suits your paper. Make sure to look at the "Author's Guide" so you can format your paper according to the guidelines for that publication. Then, submit your paper and don't get discouraged if it is not accepted right away. You may need to revise your paper and try again. To learn about the different responses you might get from journals, see our reviewer's explanation below. Did this summary help you? Yes No
- Send fan mail to authors
Reader Success Stories
RAMDEV GOHIL
Oct 16, 2017
Did this article help you?
David Okandeji
Oct 23, 2019
Revati Joshi
Feb 13, 2017
Shahzad Khan
Jul 1, 2017
Apr 7, 2017

Featured Articles

Trending Articles

Watch Articles

- Terms of Use
- Privacy Policy
- Do Not Sell or Share My Info
- Not Selling Info
Don’t miss out! Sign up for
wikiHow’s newsletter
- Privacy Policy

Home » How to Publish a Research Paper – Step by Step Guide
How to Publish a Research Paper – Step by Step Guide
Table of Contents

Publishing a research paper is an important step for researchers to disseminate their findings to a wider audience and contribute to the advancement of knowledge in their field. Whether you are a graduate student, a postdoctoral fellow, or an established researcher, publishing a paper requires careful planning, rigorous research, and clear writing. In this process, you will need to identify a research question , conduct a thorough literature review , design a methodology, analyze data, and draw conclusions. Additionally, you will need to consider the appropriate journals or conferences to submit your work to and adhere to their guidelines for formatting and submission. In this article, we will discuss some ways to publish your Research Paper.
How to Publish a Research Paper
To Publish a Research Paper follow the guide below:
- Conduct original research : Conduct thorough research on a specific topic or problem. Collect data, analyze it, and draw conclusions based on your findings.
- Write the paper : Write a detailed paper describing your research. It should include an abstract, introduction, literature review, methodology, results, discussion, and conclusion.
- Choose a suitable journal or conference : Look for a journal or conference that specializes in your research area. You can check their submission guidelines to ensure your paper meets their requirements.
- Prepare your submission: Follow the guidelines and prepare your submission, including the paper, abstract, cover letter, and any other required documents.
- Submit the paper: Submit your paper online through the journal or conference website. Make sure you meet the submission deadline.
- Peer-review process : Your paper will be reviewed by experts in the field who will provide feedback on the quality of your research, methodology, and conclusions.
- Revisions : Based on the feedback you receive, revise your paper and resubmit it.
- Acceptance : Once your paper is accepted, you will receive a notification from the journal or conference. You may need to make final revisions before the paper is published.
- Publication : Your paper will be published online or in print. You can also promote your work through social media or other channels to increase its visibility.
How to Choose Journal for Research Paper Publication
Here are some steps to follow to help you select an appropriate journal:
- Identify your research topic and audience : Your research topic and intended audience should guide your choice of journal. Identify the key journals in your field of research and read the scope and aim of the journal to determine if your paper is a good fit.
- Analyze the journal’s impact and reputation : Check the impact factor and ranking of the journal, as well as its acceptance rate and citation frequency. A high-impact journal can give your paper more visibility and credibility.
- Consider the journal’s publication policies : Look for the journal’s publication policies such as the word count limit, formatting requirements, open access options, and submission fees. Make sure that you can comply with the requirements and that the journal is in line with your publication goals.
- Look at recent publications : Review recent issues of the journal to evaluate whether your paper would fit in with the journal’s current content and style.
- Seek advice from colleagues and mentors: Ask for recommendations and suggestions from your colleagues and mentors in your field, especially those who have experience publishing in the same or similar journals.
- Be prepared to make changes : Be prepared to revise your paper according to the requirements and guidelines of the chosen journal. It is also important to be open to feedback from the editor and reviewers.
List of Journals for Research Paper Publications
There are thousands of academic journals covering various fields of research. Here are some of the most popular ones, categorized by field:
General/Multidisciplinary
- Nature: https://www.nature.com/
- Science: https://www.sciencemag.org/
- PLOS ONE: https://journals.plos.org/plosone/
- Proceedings of the National Academy of Sciences (PNAS): https://www.pnas.org/
- The Lancet: https://www.thelancet.com/
- JAMA (Journal of the American Medical Association): https://jamanetwork.com/journals/jama
Social Sciences/Humanities
- Journal of Personality and Social Psychology: https://www.apa.org/pubs/journals/psp
- Journal of Consumer Research: https://www.journals.uchicago.edu/journals/jcr
- Journal of Educational Psychology: https://www.apa.org/pubs/journals/edu
- Journal of Applied Psychology: https://www.apa.org/pubs/journals/apl
- Journal of Communication: https://academic.oup.com/joc
- American Journal of Political Science: https://ajps.org/
- Journal of International Business Studies: https://www.jibs.net/
- Journal of Marketing Research: https://www.ama.org/journal-of-marketing-research/
Natural Sciences
- Journal of Biological Chemistry: https://www.jbc.org/
- Cell: https://www.cell.com/
- Science Advances: https://advances.sciencemag.org/
- Chemical Reviews: https://pubs.acs.org/journal/chreay
- Angewandte Chemie: https://onlinelibrary.wiley.com/journal/15213765
- Physical Review Letters: https://journals.aps.org/prl/
- Journal of Geophysical Research: https://agupubs.onlinelibrary.wiley.com/journal/2156531X
- Journal of High Energy Physics: https://link.springer.com/journal/13130
Engineering/Technology
- IEEE Transactions on Neural Networks and Learning Systems: https://ieeexplore.ieee.org/xpl/RecentIssue.jsp?punumber=5962385
- IEEE Transactions on Power Systems: https://ieeexplore.ieee.org/xpl/RecentIssue.jsp?punumber=59
- IEEE Transactions on Medical Imaging: https://ieeexplore.ieee.org/xpl/RecentIssue.jsp?punumber=42
- IEEE Transactions on Control Systems Technology: https://ieeexplore.ieee.org/xpl/RecentIssue.jsp?punumber=87
- Journal of Engineering Mechanics: https://ascelibrary.org/journal/jenmdt
- Journal of Materials Science: https://www.springer.com/journal/10853
- Journal of Chemical Engineering of Japan: https://www.jstage.jst.go.jp/browse/jcej
- Journal of Mechanical Design: https://asmedigitalcollection.asme.org/mechanicaldesign
Medical/Health Sciences
- New England Journal of Medicine: https://www.nejm.org/
- The BMJ (formerly British Medical Journal): https://www.bmj.com/
- Journal of the American Medical Association (JAMA): https://jamanetwork.com/journals/jama
- Annals of Internal Medicine: https://www.acpjournals.org/journal/aim
- American Journal of Epidemiology: https://academic.oup.com/aje
- Journal of Clinical Oncology: https://ascopubs.org/journal/jco
- Journal of Infectious Diseases: https://academic.oup.com/jid
List of Conferences for Research Paper Publications
There are many conferences that accept research papers for publication. The specific conferences you should consider will depend on your field of research. Here are some suggestions for conferences in a few different fields:
Computer Science and Information Technology:
- IEEE International Conference on Computer Communications (INFOCOM): https://www.ieee-infocom.org/
- ACM SIGCOMM Conference on Data Communication: https://conferences.sigcomm.org/sigcomm/
- IEEE Symposium on Security and Privacy (SP): https://www.ieee-security.org/TC/SP/
- ACM Conference on Computer and Communications Security (CCS): https://www.sigsac.org/ccs/
- ACM Conference on Human-Computer Interaction (CHI): https://chi2022.acm.org/
Engineering:
- IEEE International Conference on Robotics and Automation (ICRA): https://www.ieee-icra.org/
- International Conference on Mechanical and Aerospace Engineering (ICMAE): http://www.icmae.org/
- International Conference on Civil and Environmental Engineering (ICCEE): http://www.iccee.org/
- International Conference on Materials Science and Engineering (ICMSE): http://www.icmse.org/
- International Conference on Energy and Power Engineering (ICEPE): http://www.icepe.org/
Natural Sciences:
- American Chemical Society National Meeting & Exposition: https://www.acs.org/content/acs/en/meetings/national-meeting.html
- American Physical Society March Meeting: https://www.aps.org/meetings/march/
- International Conference on Environmental Science and Technology (ICEST): http://www.icest.org/
- International Conference on Natural Science and Environment (ICNSE): http://www.icnse.org/
- International Conference on Life Science and Biological Engineering (LSBE): http://www.lsbe.org/
Social Sciences:
- Annual Meeting of the American Sociological Association (ASA): https://www.asanet.org/annual-meeting-2022
- International Conference on Social Science and Humanities (ICSSH): http://www.icssh.org/
- International Conference on Psychology and Behavioral Sciences (ICPBS): http://www.icpbs.org/
- International Conference on Education and Social Science (ICESS): http://www.icess.org/
- International Conference on Management and Information Science (ICMIS): http://www.icmis.org/
How to Publish a Research Paper in Journal
Publishing a research paper in a journal is a crucial step in disseminating scientific knowledge and contributing to the field. Here are the general steps to follow:
- Choose a research topic : Select a topic of your interest and identify a research question or problem that you want to investigate. Conduct a literature review to identify the gaps in the existing knowledge that your research will address.
- Conduct research : Develop a research plan and methodology to collect data and conduct experiments. Collect and analyze data to draw conclusions that address the research question.
- Write a paper: Organize your findings into a well-structured paper with clear and concise language. Your paper should include an introduction, literature review, methodology, results, discussion, and conclusion. Use academic language and provide references for your sources.
- Choose a journal: Choose a journal that is relevant to your research topic and audience. Consider factors such as impact factor, acceptance rate, and the reputation of the journal.
- Follow journal guidelines : Review the submission guidelines and formatting requirements of the journal. Follow the guidelines carefully to ensure that your paper meets the journal’s requirements.
- Submit your paper : Submit your paper to the journal through the online submission system or by email. Include a cover letter that briefly explains the significance of your research and why it is suitable for the journal.
- Wait for reviews: Your paper will be reviewed by experts in the field. Be prepared to address their comments and make revisions to your paper.
- Revise and resubmit: Make revisions to your paper based on the reviewers’ comments and resubmit it to the journal. If your paper is accepted, congratulations! If not, consider revising and submitting it to another journal.
- Address reviewer comments : Reviewers may provide comments and suggestions for revisions to your paper. Address these comments carefully and thoughtfully to improve the quality of your paper.
- Submit the final version: Once your revisions are complete, submit the final version of your paper to the journal. Be sure to follow any additional formatting guidelines and requirements provided by the journal.
- Publication : If your paper is accepted, it will be published in the journal. Some journals provide online publication while others may publish a print version. Be sure to cite your published paper in future research and communicate your findings to the scientific community.
How to Publish a Research Paper for Students
Here are some steps you can follow to publish a research paper as an Under Graduate or a High School Student:
- Select a topic: Choose a topic that is relevant and interesting to you, and that you have a good understanding of.
- Conduct research : Gather information and data on your chosen topic through research, experiments, surveys, or other means.
- Write the paper : Start with an outline, then write the introduction, methods, results, discussion, and conclusion sections of the paper. Be sure to follow any guidelines provided by your instructor or the journal you plan to submit to.
- Edit and revise: Review your paper for errors in spelling, grammar, and punctuation. Ask a peer or mentor to review your paper and provide feedback for improvement.
- Choose a journal : Look for journals that publish papers in your field of study and that are appropriate for your level of research. Some popular journals for students include PLOS ONE, Nature, and Science.
- Submit the paper: Follow the submission guidelines for the journal you choose, which typically include a cover letter, abstract, and formatting requirements. Be prepared to wait several weeks to months for a response.
- Address feedback : If your paper is accepted with revisions, address the feedback from the reviewers and resubmit your paper. If your paper is rejected, review the feedback and consider revising and resubmitting to a different journal.
How to Publish a Research Paper for Free
Publishing a research paper for free can be challenging, but it is possible. Here are some steps you can take to publish your research paper for free:
- Choose a suitable open-access journal: Look for open-access journals that are relevant to your research area. Open-access journals allow readers to access your paper without charge, so your work will be more widely available.
- Check the journal’s reputation : Before submitting your paper, ensure that the journal is reputable by checking its impact factor, publication history, and editorial board.
- Follow the submission guidelines : Every journal has specific guidelines for submitting papers. Make sure to follow these guidelines carefully to increase the chances of acceptance.
- Submit your paper : Once you have completed your research paper, submit it to the journal following their submission guidelines.
- Wait for the review process: Your paper will undergo a peer-review process, where experts in your field will evaluate your work. Be patient during this process, as it can take several weeks or even months.
- Revise your paper : If your paper is rejected, don’t be discouraged. Revise your paper based on the feedback you receive from the reviewers and submit it to another open-access journal.
- Promote your research: Once your paper is published, promote it on social media and other online platforms. This will increase the visibility of your work and help it reach a wider audience.
Journals and Conferences for Free Research Paper publications
Here are the websites of the open-access journals and conferences mentioned:
Open-Access Journals:
- PLOS ONE – https://journals.plos.org/plosone/
- BMC Research Notes – https://bmcresnotes.biomedcentral.com/
- Frontiers in… – https://www.frontiersin.org/
- Journal of Open Research Software – https://openresearchsoftware.metajnl.com/
- PeerJ – https://peerj.com/
Conferences:
- IEEE Global Communications Conference (GLOBECOM) – https://globecom2022.ieee-globecom.org/
- IEEE International Conference on Computer Communications (INFOCOM) – https://infocom2022.ieee-infocom.org/
- IEEE International Conference on Data Mining (ICDM) – https://www.ieee-icdm.org/
- ACM SIGCOMM Conference on Data Communication (SIGCOMM) – https://conferences.sigcomm.org/sigcomm/
- ACM Conference on Computer and Communications Security (CCS) – https://www.sigsac.org/ccs/CCS2022/
Importance of Research Paper Publication
Research paper publication is important for several reasons, both for individual researchers and for the scientific community as a whole. Here are some reasons why:
- Advancing scientific knowledge : Research papers provide a platform for researchers to present their findings and contribute to the body of knowledge in their field. These papers often contain novel ideas, experimental data, and analyses that can help to advance scientific understanding.
- Building a research career : Publishing research papers is an essential component of building a successful research career. Researchers are often evaluated based on the number and quality of their publications, and having a strong publication record can increase one’s chances of securing funding, tenure, or a promotion.
- Peer review and quality control: Publication in a peer-reviewed journal means that the research has been scrutinized by other experts in the field. This peer review process helps to ensure the quality and validity of the research findings.
- Recognition and visibility : Publishing a research paper can bring recognition and visibility to the researchers and their work. It can lead to invitations to speak at conferences, collaborations with other researchers, and media coverage.
- Impact on society : Research papers can have a significant impact on society by informing policy decisions, guiding clinical practice, and advancing technological innovation.
Advantages of Research Paper Publication
There are several advantages to publishing a research paper, including:
- Recognition: Publishing a research paper allows researchers to gain recognition for their work, both within their field and in the academic community as a whole. This can lead to new collaborations, invitations to conferences, and other opportunities to share their research with a wider audience.
- Career advancement : A strong publication record can be an important factor in career advancement, particularly in academia. Publishing research papers can help researchers secure funding, grants, and promotions.
- Dissemination of knowledge : Research papers are an important way to share new findings and ideas with the broader scientific community. By publishing their research, scientists can contribute to the collective body of knowledge in their field and help advance scientific understanding.
- Feedback and peer review : Publishing a research paper allows other experts in the field to provide feedback on the research, which can help improve the quality of the work and identify potential flaws or limitations. Peer review also helps ensure that research is accurate and reliable.
- Citation and impact : Published research papers can be cited by other researchers, which can help increase the impact and visibility of the research. High citation rates can also help establish a researcher’s reputation and credibility within their field.
About the author
Muhammad Hassan
Researcher, Academic Writer, Web developer
You may also like

Research Paper Introduction – Writing Guide and...

Future Research – Thesis Guide

Scope of the Research – Writing Guide and...

Chapter Summary & Overview – Writing Guide...

Research Design – Types, Methods and Examples

Theoretical Framework – Types, Examples and...
- Insights blog
Research your publishing options
Take the time to explore the journals in your field, to choose the best fit for your research. Find a journal that serves the audience you’re trying to reach, and whose aims and scope match your approach. You might also have choices to make about different publishing options, including open access.
Discover more about choosing the right journal

Draft your article
When you’ve chosen the journal you want to submit to, you’re ready to start drafting your paper. What should you be thinking about before you start writing and how can you maximize your chances of getting published?
Get advice on writing your paper
Read the instructions for authors
The instructions for authors include essential information on what you need to do to submit to your chosen journal. Follow these guidelines and you’ll know that the journal’s editorial team have everything they need to consider your article for publication.
Learn how to use the instructions for authors

Make your submission
So, you’ve done your research, chosen your target journal, written your paper, and are about to submit. Time for one final check to make sure you’ve got everything ready before heading to the journal’s submission system.
Get prepared to submit your paper

Peer review
If the journal editor thinks your article has potential for publication, they will send it out to be reviewed by two or three experts in the field. This can be a daunting prospect, but peer review is a fundamental part of getting published and can be a great opportunity to access constructive feedback on your work.
Read our comprehensive guide to peer review

Making revisions
Following peer review, you may be asked to make revisions to your article and resubmit. Take time to read through the editor and reviewers’ advice, and decide what changes you’ll make to your article. Taking their points on board will help to ensure your final article is as robust and impactful as possible.
Get guidance on revising and resubmitting

Your article is accepted
The next step is production. Copy editing begins, and we’ll contact you with your proofs. You’ll also sign a publishing agreement. If you submitted to an Open Select journal, now is the time to choose whether to publish your article open access.
Find out what happens when your article is in production

Promoting your published work
Promoting your published article is a team effort. Taylor & Francis works hard to maximize the discoverability of your work, and we’ve got lots of advice to help you share your work and amplify its impact.
Learn how to get the most from your published article

Trending topics

Boost your research career by signing up to Research Insights and receive free expert guidance on the publishing process.
Need help getting published?

Thinking about where to submit your paper? Our comprehensive guide to choosing a journal is here to help.
Free guide to choosing a journal

Call for papers in your subject area

Journals regularly ‘call for papers’, asking for submissions within a particular field or topic. Answering these is a great way to get published, making sure your research fits the journal’s aims and scope. Simply select your subject area with our handy tool to get started.
Browse upcoming events for researchers

We are delighted to share the details of our upcoming events for researchers, hosted by Taylor & Francis. Take a look at the events happening and register your place for those which are relevant to you and your research.
What do you consider important in a high-quality journal?

What factors help you decide if a journal is a good venue to publish your article, or is a reliable source of the latest research?
Let us know what is important to you in our journal quality survey.
How has our partnership with Jisc accelerated the transition to open access?

We are pleased to share a new report published by Taylor & Francis that explores the first two years of our open access partnership with the Jisc consortium. It details how the partnership has boosted the global impact of research from UK institutions.
Read our new report on how to create an enabling environment for translational research practices

How to support translational research design principles throughout the knowledge journey? Who needs to be involved?
Taylor & Francis partners with EASSH and EATRIS to explore ways to support cross disciplinary and cross sectoral collaboration.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List

How to Write and Publish a Research Paper for a Peer-Reviewed Journal
Clara busse.
1 Department of Maternal and Child Health, University of North Carolina Gillings School of Global Public Health, 135 Dauer Dr, 27599 Chapel Hill, NC USA
Ella August
2 Department of Epidemiology, University of Michigan School of Public Health, 1415 Washington Heights, Ann Arbor, MI 48109-2029 USA
Associated Data
Communicating research findings is an essential step in the research process. Often, peer-reviewed journals are the forum for such communication, yet many researchers are never taught how to write a publishable scientific paper. In this article, we explain the basic structure of a scientific paper and describe the information that should be included in each section. We also identify common pitfalls for each section and recommend strategies to avoid them. Further, we give advice about target journal selection and authorship. In the online resource 1 , we provide an example of a high-quality scientific paper, with annotations identifying the elements we describe in this article.
Electronic supplementary material
The online version of this article (10.1007/s13187-020-01751-z) contains supplementary material, which is available to authorized users.
Introduction
Writing a scientific paper is an important component of the research process, yet researchers often receive little formal training in scientific writing. This is especially true in low-resource settings. In this article, we explain why choosing a target journal is important, give advice about authorship, provide a basic structure for writing each section of a scientific paper, and describe common pitfalls and recommendations for each section. In the online resource 1 , we also include an annotated journal article that identifies the key elements and writing approaches that we detail here. Before you begin your research, make sure you have ethical clearance from all relevant ethical review boards.
Select a Target Journal Early in the Writing Process
We recommend that you select a “target journal” early in the writing process; a “target journal” is the journal to which you plan to submit your paper. Each journal has a set of core readers and you should tailor your writing to this readership. For example, if you plan to submit a manuscript about vaping during pregnancy to a pregnancy-focused journal, you will need to explain what vaping is because readers of this journal may not have a background in this topic. However, if you were to submit that same article to a tobacco journal, you would not need to provide as much background information about vaping.
Information about a journal’s core readership can be found on its website, usually in a section called “About this journal” or something similar. For example, the Journal of Cancer Education presents such information on the “Aims and Scope” page of its website, which can be found here: https://www.springer.com/journal/13187/aims-and-scope .
Peer reviewer guidelines from your target journal are an additional resource that can help you tailor your writing to the journal and provide additional advice about crafting an effective article [ 1 ]. These are not always available, but it is worth a quick web search to find out.
Identify Author Roles Early in the Process
Early in the writing process, identify authors, determine the order of authors, and discuss the responsibilities of each author. Standard author responsibilities have been identified by The International Committee of Medical Journal Editors (ICMJE) [ 2 ]. To set clear expectations about each team member’s responsibilities and prevent errors in communication, we also suggest outlining more detailed roles, such as who will draft each section of the manuscript, write the abstract, submit the paper electronically, serve as corresponding author, and write the cover letter. It is best to formalize this agreement in writing after discussing it, circulating the document to the author team for approval. We suggest creating a title page on which all authors are listed in the agreed-upon order. It may be necessary to adjust authorship roles and order during the development of the paper. If a new author order is agreed upon, be sure to update the title page in the manuscript draft.
In the case where multiple papers will result from a single study, authors should discuss who will author each paper. Additionally, authors should agree on a deadline for each paper and the lead author should take responsibility for producing an initial draft by this deadline.
Structure of the Introduction Section
The introduction section should be approximately three to five paragraphs in length. Look at examples from your target journal to decide the appropriate length. This section should include the elements shown in Fig. 1 . Begin with a general context, narrowing to the specific focus of the paper. Include five main elements: why your research is important, what is already known about the topic, the “gap” or what is not yet known about the topic, why it is important to learn the new information that your research adds, and the specific research aim(s) that your paper addresses. Your research aim should address the gap you identified. Be sure to add enough background information to enable readers to understand your study. Table Table1 1 provides common introduction section pitfalls and recommendations for addressing them.

The main elements of the introduction section of an original research article. Often, the elements overlap
Common introduction section pitfalls and recommendations
| Pitfall | Recommendation |
|---|---|
| Introduction is too generic, not written to specific readers of a designated journal. | Visit your target journal’s website and investigate the journal’s readership. If you are writing for a journal with a more general readership, like PLOS ONE, you should include more background information. A narrower journal, like the Journal of the American Mosquito Control Association, may require less background information because most of its readers have expertise in the subject matter. |
| Citations are inadequate to support claims. | If a claim could be debated, it should be supported by one or more citations. To find articles relevant to your research, consider using open-access journals, which are available for anyone to read for free. A list of open-access journals can be found here: . You can also find open-access articles using PubMed Central: |
| The research aim is vague. | Be sure that your research aim contains essential details like the setting, population/sample, study design, timing, dependent variable, and independent variables. Using such details, the reader should be able to imagine the analysis you have conducted. |
Methods Section
The purpose of the methods section is twofold: to explain how the study was done in enough detail to enable its replication and to provide enough contextual detail to enable readers to understand and interpret the results. In general, the essential elements of a methods section are the following: a description of the setting and participants, the study design and timing, the recruitment and sampling, the data collection process, the dataset, the dependent and independent variables, the covariates, the analytic approach for each research objective, and the ethical approval. The hallmark of an exemplary methods section is the justification of why each method was used. Table Table2 2 provides common methods section pitfalls and recommendations for addressing them.
Common methods section pitfalls and recommendations
| Pitfall | Recommendation |
|---|---|
| The author only describes methods for one study aim, or part of an aim. |
Be sure to check that the methods describe all aspects of the study reported in the manuscript. |
| There is not enough (or any) justification for the methods used. | You must justify your choice of methods because it greatly impacts the interpretation of results. State the methods you used and then defend those decisions. For example, justify why you chose to include the measurements, covariates, and statistical approaches. |
Results Section
The focus of the results section should be associations, or lack thereof, rather than statistical tests. Two considerations should guide your writing here. First, the results should present answers to each part of the research aim. Second, return to the methods section to ensure that the analysis and variables for each result have been explained.
Begin the results section by describing the number of participants in the final sample and details such as the number who were approached to participate, the proportion who were eligible and who enrolled, and the number of participants who dropped out. The next part of the results should describe the participant characteristics. After that, you may organize your results by the aim or by putting the most exciting results first. Do not forget to report your non-significant associations. These are still findings.
Tables and figures capture the reader’s attention and efficiently communicate your main findings [ 3 ]. Each table and figure should have a clear message and should complement, rather than repeat, the text. Tables and figures should communicate all salient details necessary for a reader to understand the findings without consulting the text. Include information on comparisons and tests, as well as information about the sample and timing of the study in the title, legend, or in a footnote. Note that figures are often more visually interesting than tables, so if it is feasible to make a figure, make a figure. To avoid confusing the reader, either avoid abbreviations in tables and figures, or define them in a footnote. Note that there should not be citations in the results section and you should not interpret results here. Table Table3 3 provides common results section pitfalls and recommendations for addressing them.
Common results section pitfalls and recommendations
| Pitfall | Recommendation |
|---|---|
| The text focuses on statistical tests rather than associations. | The relationships between independent and dependent variables are at the heart of scientific studies and statistical tests are a set of strategies used to elucidate such relationships. For example, instead of reporting that “the odds ratio is 3.4,” report that “women with exposure X were 3.4 times more likely to have disease Y.” There are several ways to express such associations, but all successful approaches focus on the relationships between the variables. |
| Causal words like “cause” and “impact” are used inappropriately | Only some study designs and analytic approaches enable researchers to make causal claims. Before you use the word “cause,” consider whether this is justified given your design. Words like “associated” or “related” may be more appropriate. |
| The direction of association unclear. |
Instead of “X is associated with Y,” say “an increase in variable X is associated with a decrease in variable Y,” a sentence which more fully describes the relationship between the two variables. |
Discussion Section
Opposite the introduction section, the discussion should take the form of a right-side-up triangle beginning with interpretation of your results and moving to general implications (Fig. 2 ). This section typically begins with a restatement of the main findings, which can usually be accomplished with a few carefully-crafted sentences.

Major elements of the discussion section of an original research article. Often, the elements overlap
Next, interpret the meaning or explain the significance of your results, lifting the reader’s gaze from the study’s specific findings to more general applications. Then, compare these study findings with other research. Are these findings in agreement or disagreement with those from other studies? Does this study impart additional nuance to well-accepted theories? Situate your findings within the broader context of scientific literature, then explain the pathways or mechanisms that might give rise to, or explain, the results.
Journals vary in their approach to strengths and limitations sections: some are embedded paragraphs within the discussion section, while some mandate separate section headings. Keep in mind that every study has strengths and limitations. Candidly reporting yours helps readers to correctly interpret your research findings.
The next element of the discussion is a summary of the potential impacts and applications of the research. Should these results be used to optimally design an intervention? Does the work have implications for clinical protocols or public policy? These considerations will help the reader to further grasp the possible impacts of the presented work.
Finally, the discussion should conclude with specific suggestions for future work. Here, you have an opportunity to illuminate specific gaps in the literature that compel further study. Avoid the phrase “future research is necessary” because the recommendation is too general to be helpful to readers. Instead, provide substantive and specific recommendations for future studies. Table Table4 4 provides common discussion section pitfalls and recommendations for addressing them.
Common discussion section pitfalls and recommendations
| Pitfall | Recommendation |
|---|---|
| The author repeats detailed results or presents new results in the discussion section. | Recall from Fig. that the discussion section should take the shape of a triangle as it moves from a specific restatement of the main findings to a broader discussion of the scientific literature and implications of the study. Specific values should not be repeated in the discussion. It is also not appropriate to include new results in the discussion section. |
| The author fails to describe the implication of the study’s limitations. | No matter how well-conducted and thoughtful, all studies have limitations. Candidly describe how the limitations affect the application of the findings. |
| Statements about future research are too generic. | Is the relationship between exposure and outcome not well-described in a population that is severely impacted? Or might there be another variable that modifies the relationship between exposure and outcome? This is your opportunity to suggest areas requiring further study in your field, steering scientific inquiry toward the most meaningful questions. |
Follow the Journal’s Author Guidelines
After you select a target journal, identify the journal’s author guidelines to guide the formatting of your manuscript and references. Author guidelines will often (but not always) include instructions for titles, cover letters, and other components of a manuscript submission. Read the guidelines carefully. If you do not follow the guidelines, your article will be sent back to you.
Finally, do not submit your paper to more than one journal at a time. Even if this is not explicitly stated in the author guidelines of your target journal, it is considered inappropriate and unprofessional.
Your title should invite readers to continue reading beyond the first page [ 4 , 5 ]. It should be informative and interesting. Consider describing the independent and dependent variables, the population and setting, the study design, the timing, and even the main result in your title. Because the focus of the paper can change as you write and revise, we recommend you wait until you have finished writing your paper before composing the title.
Be sure that the title is useful for potential readers searching for your topic. The keywords you select should complement those in your title to maximize the likelihood that a researcher will find your paper through a database search. Avoid using abbreviations in your title unless they are very well known, such as SNP, because it is more likely that someone will use a complete word rather than an abbreviation as a search term to help readers find your paper.
After you have written a complete draft, use the checklist (Fig. (Fig.3) 3 ) below to guide your revisions and editing. Additional resources are available on writing the abstract and citing references [ 5 ]. When you feel that your work is ready, ask a trusted colleague or two to read the work and provide informal feedback. The box below provides a checklist that summarizes the key points offered in this article.

Checklist for manuscript quality
(PDF 362 kb)
Acknowledgments
Ella August is grateful to the Sustainable Sciences Institute for mentoring her in training researchers on writing and publishing their research.
Code Availability
Not applicable.
Data Availability
Compliance with ethical standards.
The authors declare that they have no conflict of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
How to Write and Publish a Research Paper for a Peer-Reviewed Journal
- Open access
- Published: 30 April 2020
- Volume 36 , pages 909–913, ( 2021 )
Cite this article
You have full access to this open access article

- Clara Busse ORCID: orcid.org/0000-0002-0178-1000 1 &
- Ella August ORCID: orcid.org/0000-0001-5151-1036 1 , 2
281k Accesses
15 Citations
709 Altmetric
Explore all metrics
Communicating research findings is an essential step in the research process. Often, peer-reviewed journals are the forum for such communication, yet many researchers are never taught how to write a publishable scientific paper. In this article, we explain the basic structure of a scientific paper and describe the information that should be included in each section. We also identify common pitfalls for each section and recommend strategies to avoid them. Further, we give advice about target journal selection and authorship. In the online resource 1 , we provide an example of a high-quality scientific paper, with annotations identifying the elements we describe in this article.
Similar content being viewed by others

How to Choose the Right Journal

The Point Is…to Publish?

Writing and publishing a scientific paper
Explore related subjects.
- Artificial Intelligence
Avoid common mistakes on your manuscript.
Introduction
Writing a scientific paper is an important component of the research process, yet researchers often receive little formal training in scientific writing. This is especially true in low-resource settings. In this article, we explain why choosing a target journal is important, give advice about authorship, provide a basic structure for writing each section of a scientific paper, and describe common pitfalls and recommendations for each section. In the online resource 1 , we also include an annotated journal article that identifies the key elements and writing approaches that we detail here. Before you begin your research, make sure you have ethical clearance from all relevant ethical review boards.
Select a Target Journal Early in the Writing Process
We recommend that you select a “target journal” early in the writing process; a “target journal” is the journal to which you plan to submit your paper. Each journal has a set of core readers and you should tailor your writing to this readership. For example, if you plan to submit a manuscript about vaping during pregnancy to a pregnancy-focused journal, you will need to explain what vaping is because readers of this journal may not have a background in this topic. However, if you were to submit that same article to a tobacco journal, you would not need to provide as much background information about vaping.
Information about a journal’s core readership can be found on its website, usually in a section called “About this journal” or something similar. For example, the Journal of Cancer Education presents such information on the “Aims and Scope” page of its website, which can be found here: https://www.springer.com/journal/13187/aims-and-scope .
Peer reviewer guidelines from your target journal are an additional resource that can help you tailor your writing to the journal and provide additional advice about crafting an effective article [ 1 ]. These are not always available, but it is worth a quick web search to find out.
Identify Author Roles Early in the Process
Early in the writing process, identify authors, determine the order of authors, and discuss the responsibilities of each author. Standard author responsibilities have been identified by The International Committee of Medical Journal Editors (ICMJE) [ 2 ]. To set clear expectations about each team member’s responsibilities and prevent errors in communication, we also suggest outlining more detailed roles, such as who will draft each section of the manuscript, write the abstract, submit the paper electronically, serve as corresponding author, and write the cover letter. It is best to formalize this agreement in writing after discussing it, circulating the document to the author team for approval. We suggest creating a title page on which all authors are listed in the agreed-upon order. It may be necessary to adjust authorship roles and order during the development of the paper. If a new author order is agreed upon, be sure to update the title page in the manuscript draft.
In the case where multiple papers will result from a single study, authors should discuss who will author each paper. Additionally, authors should agree on a deadline for each paper and the lead author should take responsibility for producing an initial draft by this deadline.
Structure of the Introduction Section
The introduction section should be approximately three to five paragraphs in length. Look at examples from your target journal to decide the appropriate length. This section should include the elements shown in Fig. 1 . Begin with a general context, narrowing to the specific focus of the paper. Include five main elements: why your research is important, what is already known about the topic, the “gap” or what is not yet known about the topic, why it is important to learn the new information that your research adds, and the specific research aim(s) that your paper addresses. Your research aim should address the gap you identified. Be sure to add enough background information to enable readers to understand your study. Table 1 provides common introduction section pitfalls and recommendations for addressing them.

The main elements of the introduction section of an original research article. Often, the elements overlap
Methods Section
The purpose of the methods section is twofold: to explain how the study was done in enough detail to enable its replication and to provide enough contextual detail to enable readers to understand and interpret the results. In general, the essential elements of a methods section are the following: a description of the setting and participants, the study design and timing, the recruitment and sampling, the data collection process, the dataset, the dependent and independent variables, the covariates, the analytic approach for each research objective, and the ethical approval. The hallmark of an exemplary methods section is the justification of why each method was used. Table 2 provides common methods section pitfalls and recommendations for addressing them.
Results Section
The focus of the results section should be associations, or lack thereof, rather than statistical tests. Two considerations should guide your writing here. First, the results should present answers to each part of the research aim. Second, return to the methods section to ensure that the analysis and variables for each result have been explained.
Begin the results section by describing the number of participants in the final sample and details such as the number who were approached to participate, the proportion who were eligible and who enrolled, and the number of participants who dropped out. The next part of the results should describe the participant characteristics. After that, you may organize your results by the aim or by putting the most exciting results first. Do not forget to report your non-significant associations. These are still findings.
Tables and figures capture the reader’s attention and efficiently communicate your main findings [ 3 ]. Each table and figure should have a clear message and should complement, rather than repeat, the text. Tables and figures should communicate all salient details necessary for a reader to understand the findings without consulting the text. Include information on comparisons and tests, as well as information about the sample and timing of the study in the title, legend, or in a footnote. Note that figures are often more visually interesting than tables, so if it is feasible to make a figure, make a figure. To avoid confusing the reader, either avoid abbreviations in tables and figures, or define them in a footnote. Note that there should not be citations in the results section and you should not interpret results here. Table 3 provides common results section pitfalls and recommendations for addressing them.
Discussion Section
Opposite the introduction section, the discussion should take the form of a right-side-up triangle beginning with interpretation of your results and moving to general implications (Fig. 2 ). This section typically begins with a restatement of the main findings, which can usually be accomplished with a few carefully-crafted sentences.
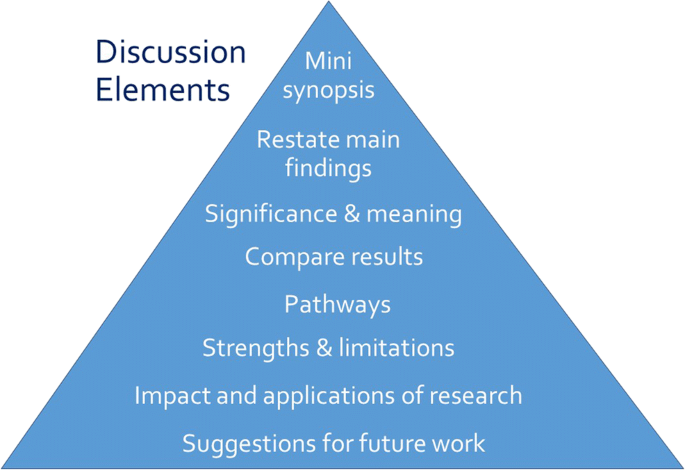
Major elements of the discussion section of an original research article. Often, the elements overlap
Next, interpret the meaning or explain the significance of your results, lifting the reader’s gaze from the study’s specific findings to more general applications. Then, compare these study findings with other research. Are these findings in agreement or disagreement with those from other studies? Does this study impart additional nuance to well-accepted theories? Situate your findings within the broader context of scientific literature, then explain the pathways or mechanisms that might give rise to, or explain, the results.
Journals vary in their approach to strengths and limitations sections: some are embedded paragraphs within the discussion section, while some mandate separate section headings. Keep in mind that every study has strengths and limitations. Candidly reporting yours helps readers to correctly interpret your research findings.
The next element of the discussion is a summary of the potential impacts and applications of the research. Should these results be used to optimally design an intervention? Does the work have implications for clinical protocols or public policy? These considerations will help the reader to further grasp the possible impacts of the presented work.
Finally, the discussion should conclude with specific suggestions for future work. Here, you have an opportunity to illuminate specific gaps in the literature that compel further study. Avoid the phrase “future research is necessary” because the recommendation is too general to be helpful to readers. Instead, provide substantive and specific recommendations for future studies. Table 4 provides common discussion section pitfalls and recommendations for addressing them.
Follow the Journal’s Author Guidelines
After you select a target journal, identify the journal’s author guidelines to guide the formatting of your manuscript and references. Author guidelines will often (but not always) include instructions for titles, cover letters, and other components of a manuscript submission. Read the guidelines carefully. If you do not follow the guidelines, your article will be sent back to you.
Finally, do not submit your paper to more than one journal at a time. Even if this is not explicitly stated in the author guidelines of your target journal, it is considered inappropriate and unprofessional.
Your title should invite readers to continue reading beyond the first page [ 4 , 5 ]. It should be informative and interesting. Consider describing the independent and dependent variables, the population and setting, the study design, the timing, and even the main result in your title. Because the focus of the paper can change as you write and revise, we recommend you wait until you have finished writing your paper before composing the title.
Be sure that the title is useful for potential readers searching for your topic. The keywords you select should complement those in your title to maximize the likelihood that a researcher will find your paper through a database search. Avoid using abbreviations in your title unless they are very well known, such as SNP, because it is more likely that someone will use a complete word rather than an abbreviation as a search term to help readers find your paper.
After you have written a complete draft, use the checklist (Fig. 3 ) below to guide your revisions and editing. Additional resources are available on writing the abstract and citing references [ 5 ]. When you feel that your work is ready, ask a trusted colleague or two to read the work and provide informal feedback. The box below provides a checklist that summarizes the key points offered in this article.

Checklist for manuscript quality
Data Availability
Michalek AM (2014) Down the rabbit hole…advice to reviewers. J Cancer Educ 29:4–5
Article Google Scholar
International Committee of Medical Journal Editors. Defining the role of authors and contributors: who is an author? http://www.icmje.org/recommendations/browse/roles-and-responsibilities/defining-the-role-of-authosrs-and-contributors.html . Accessed 15 January, 2020
Vetto JT (2014) Short and sweet: a short course on concise medical writing. J Cancer Educ 29(1):194–195
Brett M, Kording K (2017) Ten simple rules for structuring papers. PLoS ComputBiol. https://doi.org/10.1371/journal.pcbi.1005619
Lang TA (2017) Writing a better research article. J Public Health Emerg. https://doi.org/10.21037/jphe.2017.11.06
Download references
Acknowledgments
Ella August is grateful to the Sustainable Sciences Institute for mentoring her in training researchers on writing and publishing their research.
Code Availability
Not applicable.
Author information
Authors and affiliations.
Department of Maternal and Child Health, University of North Carolina Gillings School of Global Public Health, 135 Dauer Dr, 27599, Chapel Hill, NC, USA
Clara Busse & Ella August
Department of Epidemiology, University of Michigan School of Public Health, 1415 Washington Heights, Ann Arbor, MI, 48109-2029, USA
Ella August
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Ella August .
Ethics declarations
Conflicts of interests.
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
(PDF 362 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Busse, C., August, E. How to Write and Publish a Research Paper for a Peer-Reviewed Journal. J Canc Educ 36 , 909–913 (2021). https://doi.org/10.1007/s13187-020-01751-z
Download citation
Published : 30 April 2020
Issue Date : October 2021
DOI : https://doi.org/10.1007/s13187-020-01751-z
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Manuscripts
- Scientific writing
- Find a journal
- Publish with us
- Track your research

Little-known secrets for how to get published
Advice from seasoned psychologists for those seeking to publish in a journal for the first time
By Rebecca A. Clay
January 2019, Vol 50, No. 1
Print version: page 64

- Peer Review
An academic who is trying to get a journal article published is a lot like a salmon swimming upstream, says Dana S. Dunn, PhD, a member of APA’s Board of Educational Affairs. “The most important thing is persistence,” says Dunn, a psychology professor at Moravian College in Bethlehem, Pennsylvania.
But there are ways to make the journey through the publication process (see The publication process ) easier. “The more work you do up front, the more you can ensure a good outcome,” says Dunn. Among other tasks, that means finding the right venue, crafting the best possible manuscript and not giving up when asked to revise a manuscript.
The Monitor spoke with Dunn and several other senior faculty members with extensive experience publishing articles and serving as journal editors and editorial board members. Here’s their advice.
■ Target the right journals. To find the journal that’s the best fit for your article, research the journals themselves. Check each target journal’s mission statement, ask colleagues who have published there if your work is appropriate for it and read a current issue to see the kinds of articles it contains. “If your work isn’t in line with what they publish, they will reject it out of hand and you will have wasted valuable time,” says Dunn.
Also examine the composition of a journal’s editorial board, which will offer valuable clues about the kind of work the journal values—qualitative versus quantitative research, for example, or single-experiment studies versus multiple-experiment ones. You can even shoot a short email to the editor briefly summarizing your manuscript and asking if it sounds like something he or she feels would be appropriate for the journal. “Editors are pretty good at saying yes or no,” says Dunn.
In addition, let more experienced colleagues assess the strength of your study and give you some ideas about possible venues, says Jerry Suls, PhD, a professor emeritus of psychological and brain sciences at the University of Iowa in Iowa City. Ask how strong and how novel they think your results are and whether your study has any methodological limitations. Although your colleagues may not have a completely accurate view of what journals will and won’t publish, says Suls, it’s still a good idea to get a sense of what they think about your chances.
■ Balance ambition and realism. Aim high, but be realistic about where you send your manuscript. But don’t get too hung up on trying to figure out a hierarchy of which is the “best” journal, says Rose Sokol-Chang, PhD, publisher of journals at APA. Instead, she suggests, think about what you’re trying to achieve with your article. For example, some journals have a longer history, broader focus and higher impact factor, a measure of how often papers in the journal are cited compared to how much is published in the journal. For more narrowly focused research, there are journals focused on subdisciplines that are well-respected by experts and have high impact factors. You could also try highly specialized journals appropriate to your direct area of research, which are more limited in scope and readership. Keep in mind that APA publishes its own journals as well as many affiliated journals, which vary in their levels of specialization.
What you do want to avoid is publishing in one of the increasing number of journals—often online—that aren’t peer-reviewed, says Nova Southeastern University psychology professor Linda Carter Sobell, PhD. Junior faculty may not even be aware that these journals aren’t true academic journals, says Sobell, adding that one possible outcome is that they go up for tenure only to have colleagues point out that their articles are published in nonreputable journals. Tip-offs include nonuniversity addresses or requests that authors pay to publish their work. “You can’t trust them when they say they’re peer-reviewed,” says Sobell. “That could mean the secretary looks at the manuscript when it comes in.”
To check rejection rates, go to www.apa.org/pubs/journals/statistics to get a sense of the odds for APA journals. For other journals, contact the editor, check the publication’s website or directions to contributors or see if your institution subscribes to the Cabells database, which offers information on both reputable journals and those it deems “predatory.”
Also, be sure to submit your work to just one journal at a time. Shotgunning a manuscript to multiple outlets simultaneously “is completely verboten,” says Suls.
■ Hone your manuscript. Give yourself time to write the best manuscript you can, says APA Board of Scientific Affairs (BSA) member Keith F. Widaman, PhD, distinguished professor of the Graduate Division of the Graduate School of Education at the University of California, Riverside. To make sure your writing is first-rate, study “The Elements of Style,” says Widaman, who rereads the William Strunk and E.B. White classic every year or two. Learn the difference between active and passive voices, the difference between “that” and “which,” and when to use commas. “There are times when you misportray the meaning of what you’re trying to get across if you write something poorly,” he says.
Before you start writing, draft an outline with subheads that mimic how manuscripts are organized in APA journals. “Carefully constructing a manuscript helps the reader follow your thinking,” he says. Also consult APA’s newly revised Journal Article Reporting Standards (APA Style JARS), which offer guidance on what information needs to be included in a research manuscript, whether your manuscript covers quantitative research or qualitative research ( American Psychologist , Vol. 73, No. 1, 2018), suggests Sokol-Chang.
Be sure to pay close attention to details such as spelling and footnotes, Widaman adds. “If a person does a crummy job with references, for example, what other details are they not paying attention to?” To this end, take advantage of APA Style CENTRAL , which offers manuscript templates, preformatted references drawn from APA’s PsycINFO database and more.
And polish your manuscript by asking colleagues—both in and out of your specialty area—to offer constructive criticism. Make sure you haven’t overlooked relevant citations, which will suggest to reviewers that you don’t know the literature and where your work fits in. Consider having a statistician double-check your analyses.
■ Be prepared to revise. Most manuscripts are going to be rejected, so don’t take rejection personally, says Suls. It’s also extremely rare for a journal to accept a manuscript as is. Of the thousand-plus manuscripts Suls saw as associate editor of the Journal of Personality and Social Psychology, he remembers only two that received a thumbs-up without requests for at least minor revisions. “Most papers are going to be rejected or are going to be resubmitted with revisions,” says Suls. “You’re not the only one.”
Your initial reaction to reviewers’ feedback may be disappointment or even anger. Put the comments aside for a few days while you calm down. “The first thing to realize is that reviewers are not trying to do a search-and-destroy mission; they’re trying to be helpful,” says Dunn. “Often when you read the comments with a colder eye, you realize the comments are good ones that will improve your work.”

For less extreme problems, the editor may invite you to revise the rejected paper and resubmit it or suggest that you send your manuscript elsewhere. Either way, be just as diligent in revising your manuscript as you were in writing it, says Dunn. Thank the reviewers, address every comment they made and use a detailed cover letter to explain those changes, with page numbers so the editor can easily see how you’ve addressed concerns. If reviewers disagree on a particular point and the editor hasn’t chimed in, choose which side you agree with and explain why you chose that side. And if you disagree with a comment and decide to reject that advice, explain that too.
■ Gain experience. Being on the other side of the editorial process can help give you ideas about how to better craft your own manuscripts. If you’re a graduate student or junior faculty member, ask a mentor or colleague who frequently serves as a reviewer if you can become a co-reviewer. “That’s a valuable educational experience,” says Dunn. (See “ How to Review a Manuscript ” in the May 2018 Monitor for more insights.)
If you do get a chance to review someone else’s manuscript, do it well and turn it in quickly. “If it comes back in a very timely fashion and the review is a good one, that person will be used again,” says Suls. That said, junior faculty should make sure the review process does not cut into the time they devote to doing their own research and writing.
■ Keep trying. Finally, don’t give up if your article is rejected by the first publication you send it to, says BSA member Jeffrey M. Zacks, PhD, a professor of psychological and brain sciences and radiology at Washington University in St. Louis. As long as your research is fundamentally sound, says Zacks, “there’s usually another reasonable journal you can turn around and go to.”
For a set of interactive modules on publishing journal articles, reviewing journal manuscripts and other issues of importance to early career academics and researchers, see APA’s Science Career Series at www.apa.org/career-development/courses .
Further reading
Managing Your Research Data and Documentation Berenson, K.R. APA, 2017
How to Publish High-Quality Research Joireman, J., & Van Lange, P.A.M. APA, 2015
Write It Up: Practical Strategies for Writing and Publishing Journal Articles Silvia, P.J. APA, 2015
Related Articles
- The publication process
Key takeaways
1: Find the right journal for your research.
2: Write carefully and double-check your analyses.
3: Be open to revisions.
4: Don’t give up after a rejection—rework and resubmit.
Letters to the Editor
- Send us a letter
Home → Get Published → How to Publish a Research Paper: A Step-by-Step Guide
How to Publish a Research Paper: A Step-by-Step Guide
Jordan Kruszynski
- January 4, 2024

You’re in academia.
You’re going steady.
Your research is going well and you begin to wonder: ‘ How exactly do I get a research paper published?’
If this is the question on your lips, then this step-by-step guide is the one for you. We’ll be walking you through the whole process of how to publish a research paper.
Publishing a research paper is a significant milestone for researchers and academics, as it allows you to share your findings, contribute to your field of study, and start to gain serious recognition within the wider academic community. So, want to know how to publish a research paper? By following our guide, you’ll get a firm grasp of the steps involved in this process, giving you the best chance of successfully navigating the publishing process and getting your work out there.
Understanding the Publishing Process
To begin, it’s crucial to understand that getting a research paper published is a multi-step process. From beginning to end, it could take as little as 2 months before you see your paper nestled in the pages of your chosen journal. On the other hand, it could take as long as a year .
Below, we set out the steps before going into more detail on each one. Getting a feel for these steps will help you to visualise what lies ahead, and prepare yourself for each of them in turn. It’s important to remember that you won’t actually have control over every step – in fact, some of them will be decided by people you’ll probably never meet. However, knowing which parts of the process are yours to decide will allow you to adjust your approach and attitude accordingly.
Each of the following stages will play a vital role in the eventual publication of your paper:
- Preparing Your Research Paper
- Finding the Right Journal
- Crafting a Strong Manuscript
- Navigating the Peer-Review Process
- Submitting Your Paper
- Dealing with Rejections and Revising Your Paper
Step 1: Preparing Your Research Paper
It all starts here. The quality and content of your research paper is of fundamental importance if you want to get it published. This step will be different for every researcher depending on the nature of your research, but if you haven’t yet settled on a topic, then consider the following advice:
- Choose an interesting and relevant topic that aligns with current trends in your field. If your research touches on the passions and concerns of your academic peers or wider society, it may be more likely to capture attention and get published successfully.
- Conduct a comprehensive literature review (link to lit. review article once it’s published) to identify the state of existing research and any knowledge gaps within it. Aiming to fill a clear gap in the knowledge of your field is a great way to increase the practicality of your research and improve its chances of getting published.
- Structure your paper in a clear and organised manner, including all the necessary sections such as title, abstract, introduction (link to the ‘how to write a research paper intro’ article once it’s published) , methodology, results, discussion, and conclusion.
- Adhere to the formatting guidelines provided by your target journal to ensure that your paper is accepted as viable for publishing. More on this in the next section…
Step 2: Finding the Right Journal
Understanding how to publish a research paper involves selecting the appropriate journal for your work. This step is critical for successful publication, and you should take several factors into account when deciding which journal to apply for:
- Conduct thorough research to identify journals that specialise in your field of study and have published similar research. Naturally, if you submit a piece of research in molecular genetics to a journal that specialises in geology, you won’t be likely to get very far.
- Consider factors such as the journal’s scope, impact factor, and target audience. Today there is a wide array of journals to choose from, including traditional and respected print journals, as well as numerous online, open-access endeavours. Some, like Nature , even straddle both worlds.
- Review the submission guidelines provided by the journal and ensure your paper meets all the formatting requirements and word limits. This step is key. Nature, for example, offers a highly informative series of pages that tells you everything you need to know in order to satisfy their formatting guidelines (plus more on the whole submission process).
- Note that these guidelines can differ dramatically from journal to journal, and details really do matter. You might submit an outstanding piece of research, but if it includes, for example, images in the wrong size or format, this could mean a lengthy delay to getting it published. If you get everything right first time, you’ll save yourself a lot of time and trouble, as well as strengthen your publishing chances in the first place.
Step 3: Crafting a Strong Manuscript
Crafting a strong manuscript is crucial to impress journal editors and reviewers. Look at your paper as a complete package, and ensure that all the sections tie together to deliver your findings with clarity and precision.
- Begin by creating a clear and concise title that accurately reflects the content of your paper.
- Compose an informative abstract that summarises the purpose, methodology, results, and significance of your study.
- Craft an engaging introduction (link to the research paper introduction article) that draws your reader in.
- Develop a well-structured methodology section, presenting your results effectively using tables and figures.
- Write a compelling discussion and conclusion that emphasise the significance of your findings.
Step 4: Navigating the Peer-Review Process
Once you submit your research paper to a journal, it undergoes a rigorous peer-review process to ensure its quality and validity. In peer-review, experts in your field assess your research and provide feedback and suggestions for improvement, ultimately determining whether your paper is eligible for publishing or not. You are likely to encounter several models of peer-review, based on which party – author, reviewer, or both – remains anonymous throughout the process.
When your paper undergoes the peer-review process, be prepared for constructive criticism and address the comments you receive from your reviewer thoughtfully, providing clear and concise responses to their concerns or suggestions. These could make all the difference when it comes to making your next submission.
The peer-review process can seem like a closed book at times. Check out our discussion of the issue with philosopher and academic Amna Whiston in The Research Beat podcast!

Step 5: Submitting Your Paper
As we’ve already pointed out, one of the key elements in how to publish a research paper is ensuring that you meticulously follow the journal’s submission guidelines. Strive to comply with all formatting requirements, including citation styles, font, margins, and reference structure.
Before the final submission, thoroughly proofread your paper for errors, including grammar, spelling, and any inconsistencies in your data or analysis. At this stage, consider seeking feedback from colleagues or mentors to further improve the quality of your paper.
Step 6: Dealing with Rejections and Revising Your Paper
Rejection is a common part of the publishing process, but it shouldn’t discourage you. Analyse reviewer comments objectively and focus on the constructive feedback provided. Make necessary revisions and improvements to your paper to address the concerns raised by reviewers. If needed, consider submitting your paper to a different journal that is a better fit for your research.
For more tips on how to publish your paper out there, check out this thread by Dr. Asad Naveed ( @dr_asadnaveed ) – and if you need a refresher on the basics of how to publish under the Open Access model, watch this 5-minute video from Audemic Academy !
Final Thoughts
Successfully understanding how to publish a research paper requires dedication, attention to detail, and a systematic approach. By following the advice in our guide, you can increase your chances of navigating the publishing process effectively and achieving your goal of publication.
Remember, the journey may involve revisions, peer feedback, and potential rejections, but each step is an opportunity for growth and improvement. Stay persistent, maintain a positive mindset, and continue to refine your research paper until it reaches the standards of your target journal. Your contribution to your wider discipline through published research will not only advance your career, but also add to the growing body of collective knowledge in your field. Embrace the challenges and rewards that come with the publication process, and may your research paper make a significant impact in your area of study!
Looking for inspiration for your next big paper? Head to Audemic , where you can organise and listen to all the best and latest research in your field!
Keep striving, researchers! ✨
Table of Contents
Related articles.

You’re in academia. You’re going steady. Your research is going well and you begin to wonder: ‘How exactly do I get a

Behind the Scenes: What Does a Research Assistant Do?
Have you ever wondered what goes on behind the scenes in a research lab? Does it involve acting out the whims of

How to Write a Research Paper Introduction: Hook, Line, and Sinker
Want to know how to write a research paper introduction that dazzles? Struggling to hook your reader in with your opening sentences?

Blog Podcast
Privacy policy Terms of service
Subscribe to our newsletter!
Discover more from Audemic: Access any academic research via audio
Subscribe now to keep reading and get access to the full archive.
Type your email…
Continue reading
- Create an Account
How to Get Published in a Journal: 12 Proven Strategies for Academic Success

Increase your chances of academic publishing success! Learn insider tips for crafting strong research, choosing the right journal, and navigating the peer-review process.
Introduction: Starting a PhD with a goal to publish a research paper could be both exciting and daunting. Imagine the excitement of exploring something new, only to watch it transform into your worst nightmare. Sometimes, it may seem like everything is going hunky-dory until one reaches the submission process, only to return disheartened.
As you work to publish an article, you may find this journey to be flooded with both challenges and triumphs. Sometimes, the happiness of finding new insights is often ruined by the uncertainty of peer review and multiple revisions for publication. Yes–as we said—the journey to publish an article in a journal is quite a tiring process of highs and lows.
Well, fret not, in this blog, you will learn insider tips for crafting robust research, choosing the right journal, and navigating the peer-review process.
Laying the Groundwork for Success
• Solidify Your Research Question
Determined to publish a research paper? Do you know what’s the most important part of your study? Your research question. It’s the foundation of your entire research. Therefore, it’s important to keep it clear and answerable while talking about the gap you want to bridge within your research paper.
Moreover, all the hard work you put into the research paper pays off when you catch the attention of journal editors and reviewers. But have you thought about how to achieve it? Well, for that, you must ensure that your research question is novel and directly contributes to new insights and advancements.
• Choose the Right Methodology
Before you start your research, you need to sort a few things out. The most important step is to choose the most appropriate methodology. To create well-written research, you must ensure that your methods align with the standards of your field and the nature of your research.
Let’s clarify this further.
For instance, if the quantitative methods of study are appropriate for statistical analysis, qualitative methods are suitable for studying a particular subject in depth. Thus, first understand the direction where you want to take your research. After you have decided on the right methodology, the next step is to check out the limitations of your chosen methodology, so that you can analyze both the scope and constraints beforehand.
• Support Your Claims with Evidence
Now that you have selected your methodology, the next thing is to support your claims with appropriate findings that will highlight the credibility of your research paper. To start, you should introduce data collection and analysis techniques to support your claims. From experimental data to survey results to case studies, you can choose any type of evidence to support your claims. Apart from the obvious benefits, presenting a well-analyzed dataset has the potential to enhance both the validity and reliability of your findings, eventually making your manuscript more compelling.
Finding the Perfect Match: Choosing the Right Journal
• Identify Your Target Audience
The next step is to determine the type of journal that suits your research. However, you must know that aligning the content of your paper with the right target audience is the key to success. To do that, you can consider the journal’s readership and editorial focus while aiming for the one whose audience perfectly matches your research findings. This is quite a critical part of your journey to publish an article in a journal because a well-targeted journal can increase the likelihood of your article getting read and cited while creating a meaningful impact. And, if you make the wrong choice, it could pose a serious problem.
• Research Journal Impact Factors
The Journal Impact Factor (JIF) is the average number of citations to articles in a journal. The more citations, the greater the visibility of the journal.
Imagine, your paper being published in a journal with higher JIF. The chance of your work being cited increases. But is it the only factor that counts? No, there are numerous other factors that you must keep in mind, including the journal’s scope, audience, and relevance.
• Read Recent Issues and Author Guidelines
Now that you have a few journals on your target list, the next thing is to analyze each journal carefully if you are determined to publish your article. Dive in deeper and check the recent issues of your target journals to get an idea of the style, scope, and type of articles they publish. Without doing that you won’t be able to decide if the journal articles come close to your field of interest. Moreover, this will showcase how attentive you are to detail. This step can surprisingly have a positive effect on your manuscript's chances of acceptance.
Crafting a Compelling Manuscript
• Structure Your Paper Clearly
You can’t attain success without doing the actual job. To publish a manuscript, you need to organize the contents efficiently. A well-organized manuscript with clear sections not only improves the overall layout but also makes it easier for the readers to comprehend complicated figures. Don’t forget to include the regular flow in your research paper, including Introduction, Methodology, Results, Discussion, and Conclusion. Not including these can affect your journal negatively. Sticking to this format can help you convey your research more effectively.
• Write Concisely and Effectively
Your research paper can reach a diverse audience. And not everyone has the same level of knowledge as you do. There may be circumstances when not everyone can understand the technical words you put into your research, making it difficult for them to read. So always stick to a writing style that’s easy to understand. Additionally, use active voice and strong verbs to make your writing more direct.
• Proper Citation and Referencing
Another important thing to keep in mind while writing your research paper is to consider accurate and consistent citation formatting. Now you might ask why. Well, because it is important for both the credibility and integrity of your research paper. A well-cited work saves you from committing plagiarism.
To make things easier, you should follow the journal’s style guide. Moreover, you can also leverage citation management software like Mendeley to organize your references and ensure proper formatting. This tool can save time and help maintain accuracy in your citations.
Navigating the Peer-Review Process
• Respond to Reviews Constructively
Going through the peer-review process successfully is also a very important part of your research paper. Reviews and feedback can be frustrating sometimes, but it’s essential to always respond to reviewers’ comments thoughtfully and respectfully. Address each point raised by reviewers. But keep in mind addressing the errors involves correcting any identified mistakes while defending your work means justifying your scientific decisions and interpretation wisely. Approach each comment given by the reviewer with an open mind, giving room for detailed explanations or corrections, and maintain a professional tone throughout.
• Revisions and Resubmissions
Revisions in a peer-review process are very common, so don’t be scared by doing some revisions and resubmitting your work. While doing revisions, take note of the journal’s guidelines for resubmission.
• Rejection is Not Failure
Rejection is not uncommon in academic publishing. Surprisingly, studies show that top journals like Nature and Science have rejection rates of more than 90%. So you shouldn’t be dreading the final decisions. Viewing it as an end rather than an opportunity could be your biggest mistake. Review each feedback and use it to refine your research question, methodology, or analysis. After going through the reviews carefully and making the desired changes, you are all set to resubmit your work to the same journal. Another good alternative is to target a different journal with a better fit for your work.
Additional Tips:
- We know there are times when you may disagree with your reviewers on some points, but you must keep a sense of professionalism throughout the review process.
- There may be days you may want to consult your mentors or colleagues with experience in academic publishing for advice and support.
- Thirdly, if your work has reached the point where it has been reviewed, congratulations, it’s time to celebrate because getting your work peer-reviewed is an achievement in itself.
Don’t let small challenges get to you, keep writing and contributing valuable knowledge to your field.
- Find My Rep
You are here
How to get published.
You believe your research will make a contribution to your field, and you’re ready to share it with your peers far and wide, but how do you go about getting it published, and what exactly does that involve?
If this is you, this page is a great place to start. Here you’ll find guidance to taking those first steps towards publication with confidence. From what to consider when choosing a journal, to how to submit an article and what happens next.
Getting started
Choosing the right journal for you.
Submitting your article to a Sage journal
Promoting your article
Related resources you may find useful.

How to Get Your Journal Article Published guide
Our handy guide is a quick overview covering the publishing process from preparing your article and choosing a journal, to publication (5 minute read).
View the How to Get Your Journal Article Published guide

How to Get Published webinars
Free 1 hour monthly How to Get Published webinars cover topics including writing an article, navigating the peer review process, and what exactly it means when you hear “open access.” Join fellow researchers and expert speakers live, or watch our library of recordings on a variety of topics.
Browse our webinars

Sage Perspectives blog
Looking for tips on how to make sure your article goes smoothly through the peer review process, or how to write the right title for your article?
Read our blog

Sage Campus courses
Want something a bit more in-depth? Sage Campus courses are short and interactive (around 2 hours each) and cover a range of skills, including how to get published. Your library may already subscribe to the modules, or you may want to recommend that they do. Meanwhile, you can utilize the free modules.
Explore Sage Campus

Each journal has its own Aims & Scope, so the acceptance of articles is not just about quality, but also about being a good fit. Does your work reflect the scope of the Journal? Is Open Access important to you, and does the Journal have an Open Access model available? What is the readership of the Journal, and is that readership the right audience for your work? Researching the best match for your manuscript will significantly improve your chances of being accepted.
Watch our 2 minute video

If you already know in which Sage journal you’d like to publish your work, search for it and check the manuscript submission guidelines to make sure it is a good match. Or use the Sage Journal Recommender to tell us your article title and subjects and see which journals are a potential home for your manuscript. Be prepared to adjust your manuscript to match the scope and style of the desired journal.
Find journals with the Sage Journal Recommender or browse all Sage journals

Professional presentation of your work includes a precise and clear writing style, avoiding accidental plagiarism, and formatting your article to meet the criteria of your chosen journal. All of these take time and may not be skills inherent to your field of research. Sage Author Services can help you to prepare your manuscript to comply with these and other related standards, which could significantly improve your chance of acceptance.
Visit Sage Author Services
Submitting your article to a Sage journal
You’ve identified the right journal; now you need to make sure your manuscript is the perfect fit. Following the author guidelines can be the difference between possible acceptance and rejection, so it’s definitely worth following the required guidelines. We’ve a selection of resources and guides to help:
Watch How to Get Published: Submitting Your Paper (2 minute video)
Read our Article Submission infographic , a quick reminder of essentials
Here you’ll find chapter and verse on all aspects of our Manuscript Submission Guidelines
Ready to submit? Our online Submission Checklist will help you do a final check before sending your article to us.
Each journal retains editorial independence, which means their Guidelines will vary, so do go to the home page of your chosen journal to check anything you should be aware of. You can submit your article there too.

The academic world is crowded, how can you make your article stand out? If you are active on social media platforms, telling your followers about your article is one of the simplest and most effective things you can do.

Between us, we can improve the chances of your article being found, read, downloaded and cited – of your article and you making an impact. Our tips and guidance will show you how to promote your article alongside building your academic profile.
Read our tips on how to maximize your impact

- Open access at Sage
- Top reasons to publish with Sage
- How to do research and get published webinar series
- How to get published for librarians
- Sage Author Services
- Manuscript submission guidelines
- During peer review
- During and post publication
- Help and support
- Journal Editor Gateway
- Journal Reviewer Gateway
- Ethics & Responsibility
- Sage Editorial Policies
- Publication Ethics Policies
- Sage Chinese Author Gateway 中国作者资源
- Resources Home 🏠
- Try SciSpace Copilot
- Search research papers
- Add Copilot Extension
- Try AI Detector
- Try Paraphraser
- Try Citation Generator
- April Papers
- June Papers
- July Papers

The 4-Step Guide That Will Get Your Research Published

You’ve spent months and years working on your research project, sometimes sacrificing a good night’s sleep and, often, backing out of events you really wanted to go to. Finally it’s time…to get your research published!

The scholarly publishing industry is huge and there are thousands of journals for researchers to choose from. However, given the scary high rejection rates of submission in peer-reviewed journals and the 6–12 months time taken to get published, how do you know which journal is your best bet?
Here are a few steps that you can take to significantly improve your chances of getting published:
1. Browse legit journals
As of 2015, the academic publishing market had an annual revenue of $20.5 Billion . This revenue has grown tremendously over the last two years. Consequently, this growth has given rise to a large number of predatory publishers who try to scam early-career researchers in return for getting their research published. Unfamiliar with the process of research publishing and attracted by the prospect of getting published sooner than thought, early- career researchers often fall prey to these publishers.
You can take a few measures to avoid getting scammed by these predatory journals:
- Stay wary of unsolicited calls /emails — Reputable publishers don’t make cold calls or send unsolicited emails seeking submissions. It is mainly scammers who get access researchers’ details via Google Scholar, Academia.edu etc. and then do cold reach-outs.
- Use Jeffrey Beall’s list : Jeffrey Beall built this list of predatory journals and publishers . If you find a publisher suspicious, check if their name appears on this list. If it does, be sure that you’re being mugged. Hence, stay away.
- Non-indexed journals : PubMed, JSTOR, SCOPUS, SHERPA, and DOJA ( Directory of Open Journal Access ) are some of the popular databases of authentic journals. If you are unsure about a publisher’s authenticity, check if their journal is listed on these databases.
- Non-clarity on APC ( Article Processing Charge)– Most Open Access journals charge APC. This is a definite fee about which you can find information on the journal’s website. However, predatory journals often falter while quoting APC or their websites or do not have a proper APC break-down.
Read more about identifying legitimate journals .
2. Choose the best-fit journal
Allaying your fears of being scammed by a predatory publisher is just step one towards getting your research published. The real test of your efforts starts at submission, when your paper is reviewed. This is the stage where most papers are rejected for not complying to a journals’ formatting guidelines. Each journal has its own formatting, styling and referencing guidelines. Failing to comply with these leads to rejection.
One common mistake that early-career researchers make is that they write a paper first and then decide the journal to get published in. Another mistake they make is to aim for the highest-ranked journal in their field for publication. This naturally increases the chances of rejection for first-timers.
Quality and reputation of journals matter. However, credibility of journals and getting accepted faster is of prime importance.
So a much better approach is to:
- Write a list of journals in the area of your research. You can use your university’s library search or the internet to find the journals.
- Once the list is ready, re-organize it according to the journals’ relevance and quality.
- Check if the journals on your list have published on your specific topic in the past.
- Look through your references and bibliography to see if your sources come from one or more of the journals on your list.
Together, points 3 and 4, should give you a good idea of the journals you should approach to maximize your chances of getting published.
3. Understand the submission process
As mentioned earlier, not complying with the guidelines of a journal is one of the most common reasons why research papers get rejected . Once you have decided the journal you want to publish in, visit the journal’s website and read through their guidelines. Almost all journals have a different submission process.
The guidelines of each journal tend to vary across the following details:
- Minimum and maximum length of the article
- Referencing
- Formatting (includes space, font, margin, headings etc)
- British (or Australian)/American English
- Choice of medium –electronic, hard copy, or both
Use SciSpace (Formerly Typeset) to ensure that your paper is 100% compliant to journal guidelines.
Some submission advice
While submitting your article for publication make sure that you are submitting it to only one journal at a time, as most journals would refuse to consider an article for publication if it’s considered for publication in other journals. Most publications require researchers to declare that their work is not being considered for publishing in other journals.
Some journals only accept hard copy submissions through the post, while some only accept electronic submissions (in .doc, .docx), while others may require you to submit in both formats. It is, therefore, critical to read the submission guidelines carefully on the journal’s website.
4. Write a convincing journal cover letter
The role of a cover letter is to convince an editor that your research work is worth publishing in their journal. Hence, it is highly important that you write the letter with as much sincerity as you would write your manuscript text.
Here are some tips that you can use while writing the cover letter for your journal submission:
- If possible, find out the name of the editor and address her by name. You can find out the name of the editor through the journal’s online submission system. This information is generally public.
- In the first and the second paragraph of the cover letter state the name of your manuscript, include the names of the author/s, describe the reason behind your interest in the research work you have done, and the major findings from your research. Additionally, you can refer to prior work or the previous articles that you have published.
- In the next paragraph, address the aim and scope of the journal. Write how your work contributes to the aim of the journal and falls within the scope of their scientific coverage. Also mention why your work would be valuable for their readers.
- Finally, conclude the letter with statements that tell the editor that your manuscript is original and that no part of it is under consideration for publication elsewhere. A few journals also seek researchers to submit a list of the reviewers to whom your article can be sent for review. If the journal requests so, you should include the list in the concluding part the letter.
Once submitted, peer-review can take as long as six months. This primarily depends on how a publication has set up its peer-review process. A few publications have a two-stage review process wherein an editor first reviews articles to decide if they are worthy of peer-review. If your article passes this test, it’s then sent to a reviewer or a group of reviewers (these are academics from the field that you have written your article in). This process can take several months and you would, finally, get an email or a letter from the journal stating their decision.
If the journal decides to not publish your article, you would get the reviewer’s report and comments on your work. If you don’t, you can request to get them. This will help you improve your article before you send it to another journal for consideration.
It’s rare that a researcher’s work is accepted in the first attempt. However, most of the time it is not their research work, but the neglect researchers show while approaching publishers and presenting their research that fails them. If you perfect the approach you use to reach editors, you may get your research published in your first attempt!
In light of the fact that you are on the lookout for platforms that simplify research workflows, we recommend you check out SciSpace discover . Our suite of products can make your research workflows easier so that you can focus more on advancing science.

The best-in-class solution takes care of everything from literature search and discovery, profile management, research writing and formatting, and so much more.
More Stories like this
- How to write a research paper abstract?
- Top reasons for research paper rejection
- 4 Common Research Writing Mistakes (and How to Fix Them)
You might also like

Consensus GPT vs. SciSpace GPT: Choose the Best GPT for Research

Literature Review and Theoretical Framework: Understanding the Differences

Types of Essays in Academic Writing - Quick Guide (2024)
- Advanced search
- Peer review

Discover relevant research today

Advance your research field in the open

Reach new audiences and maximize your readership
ScienceOpen puts your research in the context of
Publications
For Publishers
ScienceOpen offers content hosting, context building and marketing services for publishers. See our tailored offerings
- For academic publishers to promote journals and interdisciplinary collections
- For open access journals to host journal content in an interactive environment
- For university library publishing to develop new open access paradigms for their scholars
- For scholarly societies to promote content with interactive features
For Institutions
ScienceOpen offers state-of-the-art technology and a range of solutions and services
- For faculties and research groups to promote and share your work
- For research institutes to build up your own branding for OA publications
- For funders to develop new open access publishing paradigms
- For university libraries to create an independent OA publishing environment
For Researchers
Make an impact and build your research profile in the open with ScienceOpen
- Search and discover relevant research in over 95 million Open Access articles and article records
- Share your expertise and get credit by publicly reviewing any article
- Publish your poster or preprint and track usage and impact with article- and author-level metrics
- Create a topical Collection to advance your research field
Create a Journal powered by ScienceOpen
Launching a new open access journal or an open access press? ScienceOpen now provides full end-to-end open access publishing solutions – embedded within our smart interactive discovery environment. A modular approach allows open access publishers to pick and choose among a range of services and design the platform that fits their goals and budget.
Continue reading “Create a Journal powered by ScienceOpen”
What can a Researcher do on ScienceOpen?
ScienceOpen provides researchers with a wide range of tools to support their research – all for free. Here is a short checklist to make sure you are getting the most of the technological infrastructure and content that we have to offer. What can a researcher do on ScienceOpen? Continue reading “What can a Researcher do on ScienceOpen?”
ScienceOpen on the Road
Upcoming events.
- 15 June – Scheduled Server Maintenance, 13:00 – 01:00 CEST
Past Events
- 20 – 22 February – ResearcherToReader Conference
- 09 November – Webinar for the Discoverability of African Research
- 26 – 27 October – Attending the Workshop on Open Citations and Open Scholarly Metadata
- 18 – 22 October – ScienceOpen at Frankfurt Book Fair.
- 27 – 29 September – Attending OA Tage, Berlin .
- 25 – 27 September – ScienceOpen at Open Science Fair
- 19 – 21 September – OASPA 2023 Annual Conference .
- 22 – 24 May – ScienceOpen sponsoring Pint of Science, Berlin.
- 16-17 May – ScienceOpen at 3rd AEUP Conference.
- 20 – 21 April – ScienceOpen attending Scaling Small: Community-Owned Futures for Open Access Books .
What is ScienceOpen?
- Smart search and discovery within an interactive interface
- Researcher promotion and ORCID integration
- Open evaluation with article reviews and Collections
- Business model based on providing services to publishers
Live Twitter stream
Some of our partners:.

Where to Publish a Research Paper In High School: 18 Journals and Conferences to Consider

By Alex Yang
Graduate student at Southern Methodist University
9 minute read
So you've been working super hard writing a research paper , and you’ve finally finished. Congrats! It’s a very impressive accolade already, but you might still be wondering how to publish a research paper for students. As we’ve talked about before in our Polygence blog, “ Showcasing your work and sharing it with the world is the intellectual version of ‘pics or it didn’t happen.’ ” Of course, there are lot of different ways to showcase your work , from creating a YouTube video to making a podcast. But one of the most popular ways to showcase your research is to publish your research. Publishing your research can take the great work you’ve already done and add credibility to it, and will make a stronger impression than unpublished research. Further, the process of having your work reviewed by advanced degree researchers can be a valuable experience in itself. You can receive feedback from experts and learn how to improve upon the work you’ve already done.
Before we dive into the various reputable journals and conferences to publish your work, let’s distinguish between the various academic publishing options that you have as a high school student, as there are some nuances. Quick disclaimer: this article focuses on journals and conferences as ways to showcase your work. There are also competitions where you can submit your work, and we have written guides on competing in premier competitions like Regeneron STS and competing in Regeneron ISEF .
Publishing Options for High School Students
Peer-reviewed journals.
This is rather self-explanatory, but these journals go through the peer review process, where author(s) submit their work to the journal, and the journal's editors send the work to a group of independent experts (typically grad students or other scientists with advanced degrees) in the same field or discipline. These experts are peer reviewers, who evaluate the work based on a set of predetermined criteria, including the quality of the research, the validity of the methodology, the accuracy of the data, and the originality of the findings. The peer reviewers may suggest revisions or leave comments, but ultimately the editors will decide which suggestions to give to the student.
Once you’ve received suggestions, you have the opportunity to make revisions before submitting your final product back to the journal. The editor then decides whether or not your work is published.
Non-Peer-Reviewed Journals
These are just journals that do not undergo a review process. In general, peer-reviewed journals may be seen as more credible and prestigious. However, non-peer-reviewed journals may make it easier and faster to publish your work, which can be helpful if you are pressed for time and applying to colleges soon .
Pre Print Archives
Preprint archives or servers are online repositories where student researchers can upload and share their research papers without undergoing any review process. Preprints allow students to share their findings quickly and get feedback from the scientific community, which can help improve the research while you’re waiting to hear back from journals, which typically have longer timelines and can take up to several months to publish research. Sharing your work in a preprint archive does not prohibit you from, or interfere with submitting the same work to a journal afterwards.
Research Conferences
Prefer to present your research in a presentation or verbal format? Conferences can be a great way to “publish” your research, showcase your public speaking skills, speak directly to your audience, and network with other researchers in your field.
Student-led Journals vs Graduate Student / Professor-led Journals
Some student-led journals may have peer-review, but the actual people peer-reviewing your work may be high school students. Other journals will have graduate students, PhD students, or even faculty reviewing your work. As you can imagine, there are tradeoffs to either option. With an advanced degree student reviewing your work, you can likely expect better and more accurate feedback. Plus, it’s cool to have an expert look over your work! However, this may also mean that the journal is more selective, whereas student-led journals may be easier to publish in. Nonetheless, getting feedback from anyone who’s knowledgeable can be a great way to polish your research and writing.
Strategy for Submitting to Multiple Journals
Ultimately, your paper can only be published in one peer-reviewed journal. Submitting the same paper to multiple peer-reviewed journals at the same time is not allowed, and doing so may impact its publication at any peer-reviewed journal. If your work is not accepted at one journal, however, then you are free to submit that work to your next choice and so on. Therefore, it is best to submit to journals with a strategy in mind. Consider: what journal do I ideally want to be published in? What are some back-ups if I don’t get published in my ideal journal? Preprints, like arXiv and the Research Archive of Rising Scholars, are possible places to submit your work in advance of seeking peer-reviewed publication. These are places to “stake your claim” in a research area and get feedback from the community prior to submitting your paper to its final home in a peer-reviewed journal. You can submit your work to a preprint prior to submitting at a peer-reviewed journal. However, bioRxiv, a reputable preprint server, recommends on their website that a preprint only be posted on one server, so that’s something to keep in mind as well.
Citation and Paper Formats
All of the journals listed below have specific ways that they’d like you to cite your sources, varying from styles like MLA to APA, and it’s important that you double-check the journal’s requirements for citations, titling your paper, writing your abstract, etc. Most journal websites have very detailed guides for how they want you to format your paper, so follow those closely to avoid having to wait to hear back and then resubmit your paper. If you’re looking for more guidance on citations and bibliographies check out our blog post!
18 Journals and Conferences to Publish Your Research as a High Schooler
Now that we’ve distinguished the differences between certain journals and conferences, let’s jump into some of our favorite ones. We’ve divided up our selections based on prestige and reliability, and we’ve made these selections using our experience with helping Polygence students showcase their research .
Most Prestigious Journals
Concord review.
Cost: $70 to Submit and $200 Publication Cost (if accepted)
Deadline: Fixed Deadlines in Feb 1 (Summer Issue), May 1 (Fall), August 1 (Winter), and November 1 (Spring)
Subject area: History / Social Sciences
Type of research: All types of academic articles
The Concord Review is a quarterly journal that publishes exceptional essays written by high school students on historical topics. The journal has been around since 1987 and has a great reputation, with many student winners going to great universities. Further, if your paper is published, your essays will be sent to subscribers and teachers all around the world, which is an incredible achievement.
Papers submitted tend to be around 8,000 words, so there is definitely a lot of writing involved, and the Concord Review themselves say that they are very selective, publishing only about 5% of the essays they receive.
We’ve posted our complete guide on publishing in the Concord Review here.
Journal of Emerging Investigators (JEI)
Deadline: Rolling
Subject area: STEM
Type of research: Original hypothesis-driven scientific research
JEI is an open-access publication that features scientific research papers written by middle and high school students in the fields of biological and physical sciences. The journal includes a comprehensive peer-review process, where graduate students and other professional scientists with advanced degrees will review the manuscripts and provide suggestions to improve both the project and manuscript itself. You can expect to receive feedback in 6-8 weeks.
This should be the go-to option for students that are doing hypothesis-driven, original research or research that involves original analyses of existing data (meta-analysis, analyzing publicly available datasets, etc.). This is not an appropriate fit for students writing literature reviews. Finally, a mentor or parent must submit on behalf of the student.
We’ve had many Polygence students successfully submit to JEI. Check out Hana’s research on invasive species and their effects in drought times.
STEM Fellowship Journal (SFJ)
Cost: $400 publication fee
Subject area: All Scientific Disciplines
Type of research: Conference Proceedings, Review Articles, Viewpoint Articles, Original Research
SFJ is a peer-reviewed journal published by Canadian Science Publishing that serves as a platform for scholarly research conducted by high school and university students in the STEM fields. Peer review is conducted by undergraduate, graduate student, and professional reviewers.
Depending on the kind of research article you choose to submit, SFJ provides very specific guidelines on what to include and word limits.
Other Great Journal Options
National high school journal of science (nhsjs).
Cost: $250 for publication
Deadline: Rolling
Subject area: All science disciplines
Type of research: Original research, literature review
NHSJS is a journal peer reviewed by high schoolers from around the world, with an advisory board of adult academics. Topics are STEM related, and submission types can vary from original research papers to shorter articles.
Curieux Academic Journal
Cost: $185-215
Subject area: Engineering, Humanities, and Natural Science, Mathematics, and Social Science
Type of research: Including but not limited to research papers, review articles, and humanity/social science pieces.
Curieux Academic Journal is a non-profit run by students and was founded in 2017 to publish outstanding research by high school and middle school students. Curieux publishes one issue per month (twelve per year), so there are many opportunities to get your research published.
The Young Scientists Journal
Deadline: December
Subject area: Sciences
Type of research: Original research, literature review, blog post
The Young Scientists Journal , while a popular option for students previously, has paused submissions to process a backlog. The journal is an international peer-reviewed journal run by students, and creates print issues twice a year.
The journal has also been around for a decade and has a clear track record of producing alumni who go on to work in STEM.
Here’s an example of research submitted by Polygence student Ryan to the journal.
Journal of Research High School (JRHS)
Subject area: Any academic subject including the sciences and humanities
Type of research: Original research and significant literature reviews.
JRHS is an online research journal edited by volunteer professional scientists, researchers, teachers, and professors. JRHS accepts original research and significant literature reviews in Engineering, Humanities, Natural Science, Math, and Social Sciences.
From our experience working with our students to help publish their research, this journal is currently operating with a 15-20 week turnaround time for review. This is a bit on the longer side, so be mindful of this turnaround time if you’re looking to get your work published soon.
Youth Medical Journal
Deadline: March (currently closed)
Subject area: Medical or scientific topics
Type of research: Original research, review article, blog post, magazine article
The Youth Medical Journal is an international, student-run team of 40 students looking to share medical research.
We’ve found that this journal is a good entry point for students new to research papers, but when submissions are busy, in the past they have paused submissions.
Journal of High School Science (JHSS)
Subject area: All topics
Type of research: Original research, literature review, technical notes, opinion pieces
This peer-reviewed STEAM journal publishes quarterly, with advanced degree doctors who sit on the journal’s editorial board. In addition to typical STEM subjects, the journal also accepts manuscripts related to music and theater, which is explicitly stated on their website.
Due to the current large volume of submissions, the review process takes a minimum of 4 weeks from the time of submission.
Whitman Journal of Psychology
Subject area: Psychology
Type of research: Original research, podcasts
The WWJOP is a publication run entirely by students, where research and literature reviews in the field of psychology are recognized. The journal is run out of a high school with a teacher supervisor and student staff.
The WWJOP uniquely also accepts podcast submissions, so if that’s your preferred format for showcasing your work, then this could be the journal for you!
Cost: $180 submission fee
Subject area: Humanities
Type of research: Essay submission
The Schola is a peer-reviewed quarterly journal that showcases essays on various humanities and social sciences topics authored by high school students worldwide. They feature a diverse range of subjects such as philosophy, history, art history, English, economics, public policy, and sociology.
Editors at Schola are academics who teach and do research in the humanities and social sciences
Critical Debates in Humanities, Science and Global Justice
Cost: $10 author fee
Subject area: Ethics and frontiers of science, Biology and ecosystems, Technology and Innovation, Medical research and disease, Peace and civil society, Global citizenship, identity and democracy, Structural violence and society, Psychology, Education, AI, Sociology, Computer Science, Neuroscience, Cultural politics, Politics and Justice, Computer science and math as related to policy, Public policy, Human rights, Language, Identity and Culture, Art and activism
Critical Debates is an international academic journal for critical discourse in humanities, science and contemporary global issues for emerging young scholars
International Youth Neuroscience Association Journal
Subject area: Neuroscience
Type of research: Research papers
Although this student peer-reviewed journal is not currently accepting submissions, we’ve had students recently publish here.
Here’s an example of Nevenka’s research that was published in the November 2022 issue of the journal.
Preprint Archives to Share Your Work In
Subject area: STEM, Quantitative Finance, Economics
arXiv is an open access archive supported by Cornell University, where more than 2 million scholarly articles in a wide variety of topics have been compiled. arXiv articles are not peer-reviewed, so you will not receive any feedback on your work from experts. However, your article does go through a moderation process where your work is classified into a topic area and checked for scholarly value. This process is rather quick however and according to arXiv you can expect your article to be available on the website in about 6 hours.
Although there’s no peer review process, that means the submission standards are not as rigorous and you can get your article posted very quickly, so submitting to arXiv or other preprint archives can be something you do before trying to get published in a journal.
One slight inconvenience of submitting to arXiv is that you must be endorsed by a current arXiv author, which can typically be a mentor or teacher or professor that you have. Here’s an example of a Polygence student submitting their work to arXiv, with Albert’s research on Hamiltonian Cycles.
Subject area: Biology
Type of research: Original research
bioRxiv is a preprint server for biology research, where again the research is not peer-reviewed but undergoes a check to make sure that the material is relevant and appropriate.
bioRxiv has a bit of a longer posting time, taking around 48 hours, but that’s still very quick. bioRxiv also allows for you to submit revised versions of your research if you decide to make changes.
Research Archive of Rising Scholars (RARS)
Subject area: STEM and Humanities
Type of research: Original research, review articles, poems, short stories, scripts
Research Archive of Rising Scholars is Polygence’s own preprint server! We were inspired by arXiv so we created a repository for articles and other creative submissions in STEM and the Humanities.
We launched RARS in 2022 and we’re excited to offer a space for budding scholars as they look to publish their work in journals. Compared to other preprint archives, RARS also accepts a wider range of submission types, including poems, short stories, and scripts.
Conferences to Participate In
Symposium of rising scholars.
Deadline: Twice a year - February and July
Polygence’s very own Symposium of Rising Scholars is a bi-annual academic conference where students present and share their research with their peers and experts. The Symposium also includes a College Admissions Panel and Keynote Speech. In our 8th edition of the Symposium this past March, we had 60 students presenting live, approximately 70 students presenting asynchronously, and over 100 audience members. The keynote speaker was Chang-rae Lee, award-winning novelist and professor at Stanford University.
We’re looking to have our 9th Symposium in Fall of 2023, and you can express your interest now. If you’re interested to see what our Polygence scholars have presented in the past for the Symposium, you can check out their scholar pages here.
Junior Science and Humanities Symposium (JSHS)
Deadline: Typically in November, so for 2024’s competition look to submit in Fall 2023
Subject area: STEM topics
JSHS is a Department of Defense sponsored program and competition that consists of first submitting a written report of your research. If your submission is selected, you’ll be able to participate in the regional symposium, where you can present in oral format or poster format. A select group from the regional symposium will then qualify for the national symposium.
One of the great things about JSHS compared to the journals mentioned above is that you’re allowed to work in teams and you don’t have to be a solo author. This can make the experience more fun for you and your teammates, and allow you to combine your strengths for your submission.
For young writers undertaking a high school research project and understanding how to publish a research paper for students is crucial. Knowing how to identify the right research question and understanding where to publish your research paper are key steps in this journey.
Related Content:
Top 8 Business Journals to Publish Your Research
Why Teens Should Attend the National Student Leadership Conference (NSLC)
How to Brainstorm Your Way to Perfect Research Topic Ideas
Top 20 Most Competitive Summer Programs for High School Students
Research Opportunities for High School Students
Want to start a project of your own?
Click below to get matched with one of our expert mentors who can help take your project off the ground!
- Find journals
- My journals
Register Sign in
Register or sign-in in order to manage your journal lists
Sign in or register to save a journal
To save a journal and create lists, you need to sign in to your Elsevier account.
Find the right journal for your research
Looking for the best journal match for your paper? Search the world's leading source of academic journals using your abstract or your keywords and other details.
Check if you're eligible for open access (OA) savings.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 14 August 2024
Nonlinear dynamics of multi-omics profiles during human aging
- Xiaotao Shen ORCID: orcid.org/0000-0002-9608-9964 1 , 2 , 3 na1 ,
- Chuchu Wang ORCID: orcid.org/0000-0003-2015-7331 4 , 5 na1 ,
- Xin Zhou ORCID: orcid.org/0000-0001-8089-4507 1 , 6 ,
- Wenyu Zhou 1 ,
- Daniel Hornburg ORCID: orcid.org/0000-0002-6618-7774 1 ,
- Si Wu 1 &
- Michael P. Snyder ORCID: orcid.org/0000-0003-0784-7987 1 , 6
Nature Aging ( 2024 ) Cite this article
206k Accesses
3356 Altmetric
Metrics details
- Biochemistry
- Systems biology
Aging is a complex process associated with nearly all diseases. Understanding the molecular changes underlying aging and identifying therapeutic targets for aging-related diseases are crucial for increasing healthspan. Although many studies have explored linear changes during aging, the prevalence of aging-related diseases and mortality risk accelerates after specific time points, indicating the importance of studying nonlinear molecular changes. In this study, we performed comprehensive multi-omics profiling on a longitudinal human cohort of 108 participants, aged between 25 years and 75 years. The participants resided in California, United States, and were tracked for a median period of 1.7 years, with a maximum follow-up duration of 6.8 years. The analysis revealed consistent nonlinear patterns in molecular markers of aging, with substantial dysregulation occurring at two major periods occurring at approximately 44 years and 60 years of chronological age. Distinct molecules and functional pathways associated with these periods were also identified, such as immune regulation and carbohydrate metabolism that shifted during the 60-year transition and cardiovascular disease, lipid and alcohol metabolism changes at the 40-year transition. Overall, this research demonstrates that functions and risks of aging-related diseases change nonlinearly across the human lifespan and provides insights into the molecular and biological pathways involved in these changes.
Similar content being viewed by others

Personal aging markers and ageotypes revealed by deep longitudinal profiling
Considerations for reproducible omics in aging research

Principal component-based clinical aging clocks identify signatures of healthy aging and targets for clinical intervention
Aging is a complex and multifactorial process of physiological changes strongly associated with various human diseases, including cardiovascular diseases (CVDs), diabetes, neurodegeneration and cancer 1 . The alterations of molecules (including transcripts, proteins, metabolites and cytokines) are critically important to understand the underlying mechanism of aging and discover potential therapeutic targets for aging-related diseases. Recently, the development of high-throughput omics technologies has enabled researchers to study molecular changes at the system level 2 . A growing number of studies have comprehensively explored the molecular changes that occur during aging using omics profiling 3 , 4 , and most focus on linear changes 5 . It is widely recognized that the occurrence of aging-related diseases does not follow a proportional increase with age. Instead, the risk of these diseases accelerates at specific points throughout the human lifespan 6 . For example, in the United States, the prevalence of CVDs (encompassing atherosclerosis, stroke and myocardial infarction) is approximately 40% between the ages of 40 and 59, increases to about 75% between 60 and 79 and reaches approximately 86% in individuals older than 80 years 7 . Similarly, also in the United States, the prevalence of neurodegenerative diseases, such as Parkinson’s disease and Alzheimer’s disease, exhibits an upward trend as well as human aging progresses, with distinct turning points occurring around the ages of 40 and 65, respectively 8 , 9 , 10 . Some studies also found that brain aging followed an accelerated decline in flies 11 and chimpanzees 12 that lived past middle age and advanced age.
The observation of a nonlinear increase in the prevalence of aging-related diseases implies that the process of human aging is not a simple linear trend. Consequently, investigating the nonlinear changes in molecules will likely reveal previously unreported molecular signatures and mechanistic insights. Some studies examined the nonlinear alterations of molecules during human aging 13 . For instance, nonlinear changes in RNA and protein expression related to aging have been documented 14 , 15 , 16 . Moreover, certain DNA methylation sites have exhibited nonlinear changes in methylation intensity during aging, following a power law pattern 17 . Li et al. 18 identified the 30s and 50s as transitional periods during women’s aging. Although aging patterns are thought to reflect the underlying biological mechanisms, the comprehensive landscape of nonlinear changes of different types of molecules during aging remains largely unexplored. Remarkably, the global monitoring of nonlinear changing molecular profiles throughout human aging has yet to be fully used to extract basic insights into the biology of aging.
In the present study, we conducted a comprehensive deep multi-omics profiling on a longitudinal human cohort comprising 108 individuals aged from 25 years to 75 years. The cohort was followed over a span of several years (median, 1.7 years), with the longest monitoring period for a single participant reaching 6.8 years (2,471 days). Various types of omics data were collected from the participants’ biological samples, including transcriptomics, proteomics, metabolomics, cytokines, clinical laboratory tests, lipidomics, stool microbiome, skin microbiome, oral microbiome and nasal microbiome. The investigation explored the changes occurring across different omics profiles during human aging. Remarkably, many molecular markers and biological pathways exhibited a nonlinear pattern throughout the aging process, thereby providing valuable insight into periods of dramatic alterations during human aging.
Most of the molecules change nonlinearly during aging
We collected longitudinal biological samples from 108 participants over several years, with a median tracking period of 1.7 years and a maximum period of 6.8 years, and conducted multi-omics profiling on the samples. The participants were sampled every 3–6 months while healthy and had diverse ethnic backgrounds and ages ranging from 25 years to 75 years (median, 55.7 years). The participants’ body mass index (BMI) ranged from 19.1 kg m −2 to 40.8 kg m −2 (median, 28.2 kg m −2 ). Among the participants, 51.9% were female (Fig. 1a and Extended Data Fig. 1a–d ). For each visit, we collected blood, stool, skin swab, oral swab and nasal swab samples. In total, 5,405 biological samples (including 1,440 blood samples, 926 stool samples, 1,116 skin swab samples, 1,001 oral swab samples and 922 nasal swab samples) were collected. The biological samples were used for multi-omics data acquisition (including transcriptomics from peripheral blood mononuclear cells (PBMCs), proteomics from plasma, metabolomics from plasma, cytokines from plasma, clinical laboratory tests from plasma, lipidomics from plasma, stool microbiome, skin microbiome, oral microbiome and nasal microbiome; Methods ). In total, 135,239 biological features (including 10,346 transcripts, 302 proteins, 814 metabolites, 66 cytokines, 51 clinical laboratory tests, 846 lipids, 52,460 gut microbiome taxons, 8,947 skin microbiome taxons, 8,947 oral microbiome taxons and 52,460 nasal microbiome taxons) were acquired, resulting in 246,507,456,400 data points (Fig. 1b and Extended Data Fig. 1e,f ). The average sampling period and number of samples for each participant were 626 days and 47 samples, respectively. Notably, one participant was deeply monitored for 6.8 years (2,471 days), during which 367 samples were collected (Fig. 1c ). Overall, this extensive and longitudinal multi-omics dataset enables us to examine the molecular changes that occur during the human aging process. The detailed characteristics of all participants are provided in the Supplementary Data . For each participant, the omics data were aggregated and averaged across all healthy samples to represent the individual’s mean value, as detailed in the Methods section. Compared to cross-sectional cohorts, which have only a one-time point sample from each participant, our longitudinal dataset, which includes multiple time point samples from each participant, is more robust for detecting complex aging-related changes in molecules and functions. This is because analysis of multi-time point samples can detect participants’ baseline and robustly evaluate individuals’ longitudinal molecular changes.
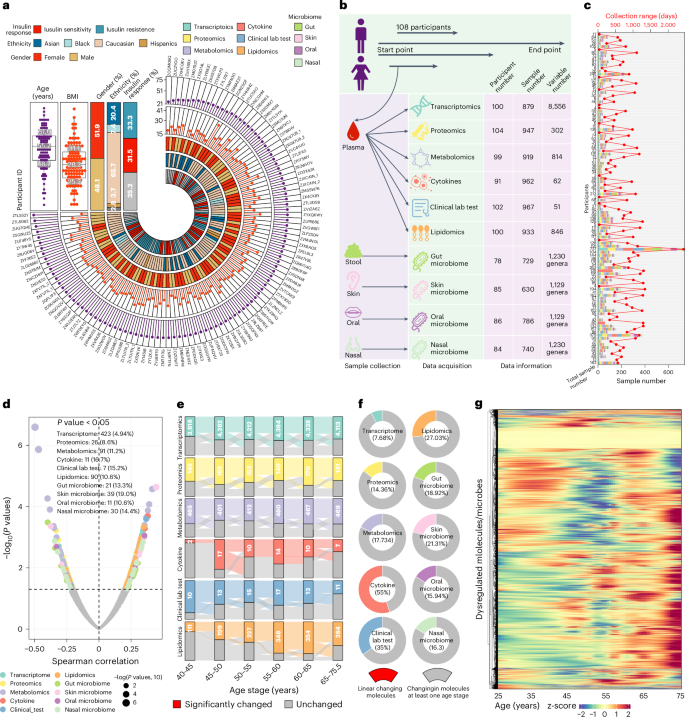
a , The demographics of the 108 participants in the study are presented. b , Sample collection and multi-omics data acquisition of the cohort. Four types of biological samples were collected, and 10 types of omics data were acquired. c , Collection time range and sample numbers for each participant. The top x axis represents the collection range for each participant (read line), and the bottom x axis represents the sample number for each participant (bar plot). Bars are color-coded by omics type. d , Significantly changed molecules and microbes during aging were detected using the Spearman correlation approach ( P < 0.05). The P values were not adjusted ( Methods ). Dots are color-coded by omics type. e , Differential expressional molecules/microbes in different age ranges compared to baseline (25–40 years old, two-sided Wilcoxon test, P < 0.05). The P values were not adjusted ( Methods ). f , The linear changing molecules comprised only a small part of dysregulated molecules in at least one age range. g , Heatmap depicting the nonlinear changing molecules and microbes during human aging.
We included samples only from healthy visits and adjusted for confounding factors (for example, BMI, sex, insulin resistance/insulin sensitivity (IRIS) and ethnicity; Extended Data Fig. 1a–d ), allowing us to discern the molecules and microbes genuinely associated with aging ( Methods ). Two common and traditional approaches, linear regression and Spearman correlation, were first used to identify the linear changing molecules during human aging 5 . The linear regression method is commonly used for linear changing molecules. As expected, both approaches have very high consistent results for each type of omics data (Supplementary Fig. 1a ). For convenience, the Spearman correlation approach was used in the analysis. Interestingly, only a small portion of all the molecules and microbes (749 out of 11,305, 6.6%; only genus level was used for microbiome data; Methods ) linearly changed during human aging (Fig. 1d and Supplementary Fig. 1b ), consistent with our previous studies 5 ( Methods ). Next, we examined nonlinear effects by categorizing all participants into distinct age stages according to their ages and investigated the dysregulated molecules within each age stage compared to the baseline (25–40 years old; Methods ). Interestingly, using this approach, 81.03% of molecules (9,106 out of 11,305) exhibited changes in at least one age stage compared to the baseline (Fig. 1e and Extended Data Fig. 2a ). Remarkably, the percentage of linear changing molecules was relatively small compared to the overall dysregulated molecules during aging (mean, 16.2%) (Fig. 1f and Extended Data Fig. 2b ). To corroborate our findings, we employed a permutation approach to calculate permutated P values, which yielded consistent results ( Methods ). The heatmap depicting all dysregulated molecules also clearly illustrates pronounced nonlinear changes (Fig. 1g ). Taken together, these findings strongly suggest that a substantial number of molecules and microbes undergo nonlinear changes throughout human aging.
Clustering reveals nonlinear multi-omics changes during aging
Next, we assessed whether the multi-omics data collected from the longitudinal cohort could serve as reliable indicators of the aging process. Our analysis revealed a substantial correlation between a significant proportion of the omics data and the ages of the participants (Fig. 2a ). Particularly noteworthy was the observation that, among all the omics data examined, metabolomics, cytokine and oral microbiome data displayed the strongest association with age (Fig. 2a and Extended Data Fig. 3a–c ). Partial least squares (PLS) regression was further used to compare the strength of the age effect across different omics data types. The results are consistent with the results presented above in Fig. 2a ( Methods ). These findings suggest the potential utility of these datasets as indicators of the aging process while acknowledging that further research is needed for validation 4 . As the omics data are not accurately matched across all the samples, we then smoothed the omics data using our previously published approach 19 ( Methods and Supplementary Fig. 2a–c ). Next, to reveal the specific patterns of molecules that change during human aging, we then grouped all the molecules with similar trajectories using an unsupervised fuzzy c-means clustering approach 19 ( Methods , Fig. 3b and Supplementary Fig. 2d,e ). We identified 11 clusters of molecular trajectories that changed during aging, which ranged in size from 638 to 1,580 molecules/microbes (Supplementary Fig. 2f and Supplementary Data ). We found that most molecular patterns exhibit nonlinear changes, indicating that aging is not a linear process (Fig. 2b ). Among the 11 identified clusters, three distinct clusters (2, 4 and 5) displayed compelling, straightforward and easily understandable patterns that spanned the entire lifespan (Fig. 2c ). Most molecules within these three clusters primarily consist of transcripts (Supplementary Fig. 2f ), which is expected because transcripts dominate the multi-omics data (8,556 out of 11,305, 75.7%). Cluster 4 exhibits a relatively stable pattern until approximately 60 years of age, after which it shows a rapid decrease (Fig. 2c ). Conversely, clusters 2 and 5 display fluctuations before 60 years of age, followed by a sharp increase and an upper inflection point at approximately 55–60 years of age (Fig. 2c ). We also attempted to observe this pattern of molecular change during aging individually. The participant with the longest follow-up period of 6.8 years (Fig. 1c ) approached the age of 60 years (range, 59.5–66.3 years; Extended Data Fig. 1g ), and it was not possible to identify obvious patterns in this short time window (Supplementary Fig. 2g ). Tracking individuals longitudinally over longer periods (decades) will be required to observe these trajectories at an individual level.
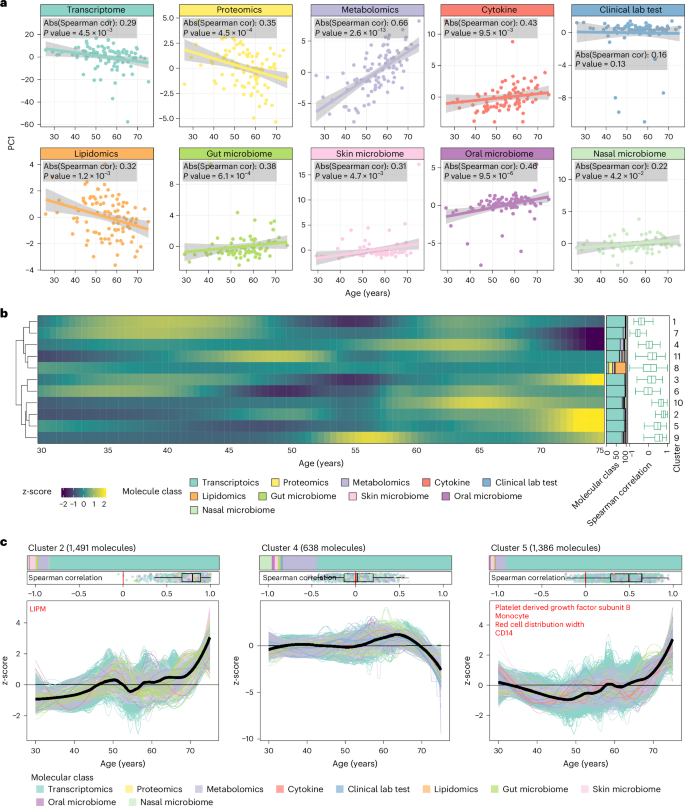
a , Spearman correlation (cor) between the first principal component and ages for each type of omics data. The shaded area around the regression line represents the 95% confidence interval. b , The heatmap shows the molecular trajectories in 11 clusters during human aging. The right stacked bar plots show the percentages of different kinds of omics data, and the right box plots show the correlation distribution between features and ages ( n = 108 participants). c , Three notable clusters of molecules that exhibit clear and straightforward nonlinear changes during human aging. The top stacked bar plots show the percentages of different kinds of omics data, and the top box plots show the correlation distribution between features and ages ( n = 108 participants). The box plot shows the median (line), interquartile range (IQR) (box) and whiskers extending to 1.5 × IQR. Bars and lines are color-coded by omics type. Abs, absolute.
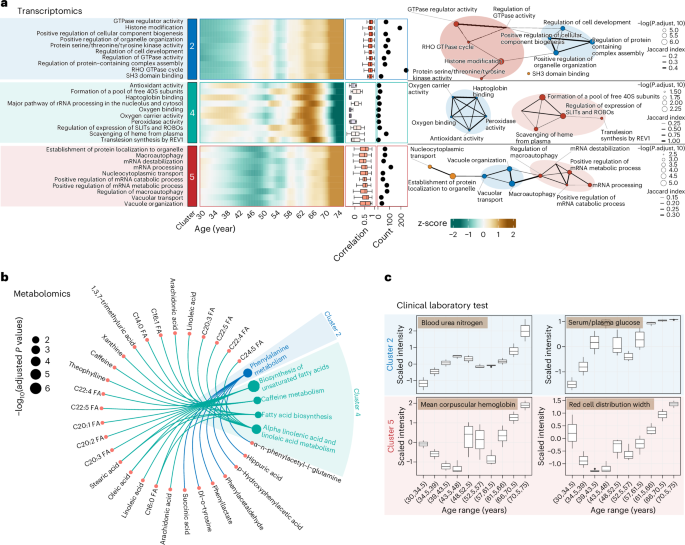
a , Pathway enrichment and module analysis for each transcriptome cluster. The left panel is the heatmap for the pathways that undergo nonlinear changes across aging. The right panel is the pathway similarity network ( Methods ) ( n = 108 participants). b , Pathway enrichment for metabolomics in each cluster. Enriched pathways and related metabolites are illustrated (Benjamini–Hochberg-adjusted P < 0.05). c , Four clinical laboratory tests that change during human aging: blood urea nitrogen, serum/plasma glucose, mean corpuscular hemoglobin and red cell distribution width ( n = 108 participants). The box plot shows the median (line), interquartile range (IQR) (box) and whiskers extending to 1.5 × IQR.
Although confounders, including sex, were corrected before analysis ( Methods ), we acknowledge that the age range for menopause in females is typically between 45 years and 55 years of age 20 , which is very close to the major transition points in all three clusters (Fig. 2c ). Therefore, we conducted further investigation into whether the menopausal status of females in the dataset contributed to the observed transition point at approximately 55 years of age (Fig. 2c ) by performing separate clustering analyses on the male and female datasets. Surprisingly, both the male and female datasets exhibited similar clusters, as illustrated in Extended Data Fig. 4a . This suggests that the transition point observed at approximately 55 years of age is not solely attributed to female menopause but, rather, represents a common phenomenon in the aging process of both sexes. This result is consistent with previous studies 14 , 15 , further supporting the notion that this transition point is a major characteristic feature of human aging. Moreover, to investigate the possibility that the transcriptomics data might skew the results toward transcriptomic changes as age-related factors, we conducted two additional clustering analyses—one focusing solely on transcriptomic data and another excluding it. Interestingly, both analyses yielded nearly identical three-cluster configurations, as observed using the complete omics dataset (Extended Data Fig. 4b ). This reinforces the robustness of the identified clusters and confirms that they are consistent across various omics platforms, not just driven by transcriptomic data.
Nonlinear changes in function and disease risk during aging
To gain further insight into the biological functions associated with the nonlinear changing molecules within the three identified clusters, we conducted separate functional analyses for transcriptomics, proteomics and metabolomics datasets for all three clusters. In brief, we constructed a similarity network using enriched pathways from various databases (Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome) and identified modules to eliminate redundant annotations. We then used all modules from different databases to reduce redundancy further using the same approach and define the final functional modules ( Methods , Extended Data Fig. 4c and Supplementary Data ). We identified some functional modules that were reported in previous studies, but we defined their more accurate patterns of change during human aging. Additionally, we also found previously unreported potential functional modules during human aging ( Supplementary Data ). For instance, in cluster 2, we identified a transcriptomic module associated with GTPase activity (adjusted P = 1.64 × 10 −6 ) and histone modification (adjusted P = 6.36 × 10 −7 ) (Fig. 3a ). Because we lack epigenomic data in this study, our findings should be validated through additional experiments in the future. GTPase activity is closely correlated with programmed cell death (apoptosis), and some previous studies showed that this activity increases during aging 21 . Additionally, histone modifications have been demonstrated to increase during human aging 22 . In cluster 4, we identified one transcriptomics module associated with oxidative stress; this module includes antioxidant activity, oxygen carrier activity, oxygen binding and peroxidase activity (adjusted P = 0.029) (Fig. 3a ). Previous studies demonstrated that oxidative stress and many reactive oxygen species (ROS) are positively associated with increased inflammation in relation to aging 23 . In cluster 5, the first transcriptomics module is associated with mRNA stability, which includes mRNA destabilization (adjusted P = 0.0032), mRNA processing (adjusted P = 3.2 × 10 −4 ), positive regulation of the mRNA catabolic process (adjusted P = 1.46 × 10 −4 ) and positive regulation of the mRNA metabolic process (adjusted P = 0.00177) (Fig. 3a ). Previous studies showed that mRNA turnover is associated with aging 24 . The second module is associated with autophagy (Fig. 3a ), which increases during human aging, as demonstrated in previous studies 25 .
In addition, we also identified certain modules in the clusters that suggest a nonlinear increase in several disease risks during human aging. For instance, in cluster 2, where components increase gradually and then rapidly after age 60, the phenylalanine metabolism pathway (adjusted P = 4.95 × 10 −4 ) was identified (Fig. 3b ). Previous studies showed that aging is associated with a progressive increase in plasma phenylalanine levels concomitant with cardiac dysfunction, and dysregulated phenylalanine catabolism is a factor that triggers deviations from healthy cardiac aging trajectories 26 . Additionally, C-X-C motif chemokine 5 (CXCL5 or ENA78) from proteomics data, which has higher concentrations in atherosclerosis 27 , is also detected in cluster 2 ( Supplementary Data ). The clinical laboratory test blood urea nitrogen, which provides important information about kidney function, is also detected in cluster 2 (Fig. 3c ). This indicates that kidney function nonlinearly decreases during aging. Furthermore, the clinical laboratory test for serum/plasma glucose, a marker of type 2 diabetes (T2D), falls within cluster 2. This is consistent with and supported by many previous studies demonstrating that aging is a major risk factor for T2D 28 . Collectively, these findings suggest a nonlinear escalation in the risk of cardiovascular and kidney diseases and T2D with advancing age, particularly after the age of 60 years (Fig. 2c ).
The identified modules in cluster 4 also indicate a nonlinear increase in disease risks. For instance, the unsaturated fatty acids biosynthesis pathway (adjusted P = 4.71 × 10 −7 ) is decreased in cluster 4. Studies have shown that unsaturated fatty acids are helpful in reducing CVD risk and maintaining brain function 29 , 30 . The pathway of alpha-linolenic acid and linolenic acid metabolism (adjusted P = 1.32 × 10 −4 ) can reduce aging-associated diseases, such as CVD 31 . We also detected the caffeine metabolism pathway (adjusted P = 7.34 × 10 −5 ) in cluster 4, which suggests that the ability to metabolize caffeine decreases during aging. Additionally, the cytokine MCP1 (chemokine (C-C motif) ligand 2 (CCL2)), a member of the CC chemokine family, plays an important immune regulatory role and is also in cluster 4 ( Supplementary Data ). These findings further support previous observations and highlight the nonlinear increase in age-related disease risk as individuals age.
Cluster 5 comprises the clinical tests of mean corpuscular hemoglobin and red cell distribution width (Fig. 3c ). These tests assess the average hemoglobin content per red blood cell and the variability in the size and volume of red blood cells, respectively. These findings align with the aforementioned transcriptomic data, which suggest a nonlinear reduction in the oxygen-carrying capacity associated with the aging process.
Aside from these three distinct clusters (Fig. 2c ), we also conducted pathway enrichment analysis across all other eight clusters, which displayed highly nonlinear trajectories, employing the same method (Fig. 2b and Supplementary Data ). Notably, cluster 11 exhibited a consistent increase up until the age of 50, followed by a decline until the age of 56, after which no substantial changes were observed up to the age of 75. A particular transcriptomics module related to DNA repair was identified, encompassing three pathways: positive regulation of double-strand break repair (adjusted P = 0.042), peptidyl−lysine acetylation (adjusted P = 1.36 × 10 −5 ) and histone acetylation (adjusted P = 3.45 × 10 −4 ) (Extended Data Fig. 4d ). These three pathways are critical in genomic stability, gene expression and metabolic balances, with their levels diminishing across the human lifespan 32 , 33 , 34 . Our findings reveal a nonlinear alteration across the human lifespan in these pathways, indicating an enhancement in DNA repair capabilities before the age of 50, a marked reduction between the ages of 50 and 56 and stabilization after that until the age of 75. The pathway enrichment results for all clusters are detailed in the Supplementary Data .
Altogether, the comprehensive functional analysis offers valuable insights into the nonlinear changes observed in molecular profiles and their correlations with biological functions and disease risks across the human lifespan. Our findings reveal that individuals aged 60 and older exhibit increased susceptibility to CVD, kidney issues and T2D. These results carry important implications for both the diagnosis and prevention of these diseases. Notably, many clinically actionable markers were identified, which have the potential for improved healthcare management and enhanced overall well-being of the aging population.
Uncovering waves of aging-related molecules during aging
Although the trajectory clustering approaches described above effectively identify nonlinear changing molecules and microbes that exhibit clear and compelling patterns throughout human aging, it may not be as effective in capturing substantial changes that occur at specific chronological aging periods. In such cases, alternative analytical approaches may be necessary to detect and characterize these dynamics. To gain a comprehensive understanding of changes in multi-omics profiling during human aging, we used a modified version of the DE-SWAN algorithm 14 , as described in the Methods section. This algorithm identifies dysregulated molecules and microbes throughout the human lifespan by analyzing molecule levels within 20-year windows and comparing two groups in 10-year parcels while sliding the window incrementally from young to old ages 14 . Using this approach and multiomics data, we detected changes at specific stages of lifespan and uncovered the sequential effects of aging. Our analysis revealed thousands of molecules exhibiting changing patterns throughout aging, forming distinct waves, as illustrated in Fig. 3a . Notably, we observed two prominent crests occurring around the ages of 45 and 65, respectively (Fig. 4a ). Notably, too, these crests were consistent with findings from a previous study that included only proteomics data 14 . Specifically, crest 2 aligns with our previous trajectory clustering result, indicating a turning point at approximately 60 years of age (Fig. 2c ).
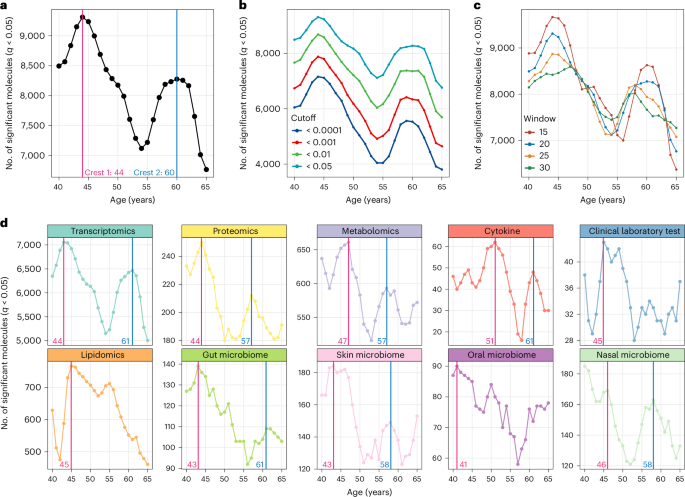
a , Number of molecules and microbes differentially expressed during aging. Two local crests at the ages of 44 years and 60 years were identified. b , c , The same waves were detected using different q value ( b ) and window ( c ) cutoffs. d , The number of molecules/microbes differentially expressed for different types of omics data during human aging.
To demonstrate the significance of the two crests, we employed different q value cutoffs and sliding window parameters, which consistently revealed the same detectable waves (Fig. 4b,c and Supplementary Fig. 4a,b ). Furthermore, when we permuted the ages of individuals, the crests disappeared (Supplementary Figs. 3a and 4c ) ( Methods ). These observations highlight the robustness of the two major waves of aging-related molecular changes across the human lifespan. Although we already accounted for confounders before our statistical analysis, we took additional steps to explore their impact. Specifically, we investigated whether confounders, such as insulin sensitivity, sex and ethnicity, differed between the two crests across various age ranges. As anticipated, these confounders did not show significant differences across other age brackets (Supplementary Fig. 4d ). This further supports our conclusion that the observed differences in the two crests are attributable to age rather than other confounding variables.
The identified crests represent notable milestones in the aging process and suggest specific age ranges where substantial molecular alterations occur. Therefore, we investigated the age-related waves for each type of omics data. Interestingly, most types of omics data exhibited two distinct crests that were highly robust (Fig. 3b and Supplementary Fig. 4 ). Notably, the proteomics data displayed two age-related crests at ages around 40 years and 60 years. Only a small overlap was observed between our dataset and the results from the previous study (1,305 proteins versus 302 proteins, with only 75 proteins overlapping). The observed pattern in our study was largely consistent with the previous findings 14 . However, our finding that many types of omics data, including transcriptomics, proteomics, metabolomics, cytokine, gut microbiome, skin microbiome and nasal microbiome, exhibit these waves, often with a similar pattern as the proteomics data (Fig. 4d ), supports the hypothesis that aging-related changes are not limited to a specific omics layer but, rather, involve a coordinated and systemic alteration across multiple molecular components. Identifying consistent crests across different omics data underscores the robustness and reliability of these molecular milestones in the aging process.
Next, we investigated the roles and functions of dysregulated molecules within two distinct crests. Notably, we found that the two crests related to aging predominantly consisted of the same molecules (Supplementary Fig. 6 ). To focus on the unique biological functions associated with each crest and eliminate commonly occurring molecules, we removed background molecules present in most stages. To explore the specific biological functions associated with each type of omics data (transcriptomics, proteomics and metabolomics) for both crests, we employed the function annotation approach described above ( Methods ). In brief, we constructed a similarity network of enriched pathways and identified modules to remove redundant annotations (Supplementary Fig. 6 and Extended Data Fig. 5a,b ). Furthermore, we applied the same approach to all modules to reduce redundancy and identify the final functional modules ( Methods and Extended Data Fig. 6a ). Our analysis revealed significant changes in multiple modules associated with the two crests (Extended Data Fig. 6b–d ). To present the results clearly, Fig. 5a displays the top 20 pathways (according to adjusted P value) for each type of omics data, and the Supplementary Data provides a comprehensive list of all identified functional modules.
a , Pathway enrichment and biological functional module analysis for crests 1 and 2. Dots and lines are color-coded by omics type. b , The overlapping of molecules between two crests and three clusters.
Interestingly, the analysis identifies many dysregulated functional modules in crests 1 and 2, indicating a nonlinear risk for aging-related diseases. In particular, several modules associated with CVD were identified in both crest 1 and crest 2 (Fig. 5a ), which is consistent with the above results (Fig. 3b ). For instance, the dysregulation of platelet degranulation (crest 1: adjusted P = 1.77 × 10 −30 ; crest 2: adjusted P = 1.73 × 10 −26 ) 35 , 36 , complement cascade (crest 1: adjusted P = 3.84 × 10 −30 ; crest 2: adjusted P = 2.02 × 10 −28 ), complement and coagulation cascades (crest 1: adjusted P = 1.78 × 10 −46 ; crest 2: adjusted P = 2.02 × 10 −28 ) 37 , 38 , protein activation cascade (crest 1: adjusted P = 1.56 × 10 −17 ; crest 2: adjusted P = 1.61 × 10 −8 ) and protease binding (crest 1: adjusted P = 2.7 × 10 −6 ; crest 2: adjusted P = 0.0114) 39 have various effects on the cardiovascular system and can contribute to various CVDs. Furthermore, blood coagulation (crest 1: adjusted P = 1.48 × 10 −28 ; crest 2: adjusted P = 9.10 × 10 −17 ) and fibrinolysis (crest 1: adjusted P = 2.11 × 10 −15 ; crest 2: adjusted P = 1.64 × 10 −10 ) were also identified, which are essential processes for maintaining blood fluidity, and dysregulation in these modules can lead to thrombotic and cardiovascular events 40 , 41 . We also identified certain dysregulated metabolic modules associated with CVD. For example, aging has been linked to an incremental rise in plasma phenylalanine levels (crest 1: adjusted P = 9.214 × 10 −4 ; crest 2: adjusted P = 0.0453), which can contribute to the development of cardiac hypertrophy, fibrosis and dysfunction 26 . Branched-chain amino acids (BCAAs), including valine, leucine and isoleucine (crest 1: adjusted P : not significant (NS); crest 2: adjusted P = 0.0173), have also been implicated in CVD development 42 , 43 and T2D, highlighting their relevance in CVD pathophysiology. Furthermore, research suggests that alpha-linolenic acid (ALA) and linoleic acid metabolism (crest 1: adjusted P : NS; crest 2: adjusted P = 0.0217) may be protective against coronary heart disease 44 , 45 . Our investigation also identified lipid metabolism modules that are associated with CVD, including high-density lipoprotein (HDL) remodeling (crest 1: adjusted P = 1.073 × 10 −8 ; crest 2: adjusted P = 2.589 × 10 −9 ) and glycerophospholipid metabolism (crest 1: adjusted P : NS; crest 2: adjusted P = 0.0033), which influence various CVDs 46 , 47 , 48 .
In addition, the dysregulation of skin and muscle stability was found to be increased at crest 1 and crest 2, as evidenced by the identification of numerous modules associated with these processes (Fig. 5a,b ). This suggests that the aging of skin and muscle is markedly accelerated at crest 1 and crest 2. The extracellular matrix (ECM) provides structural stability, mechanical strength, elasticity and hydration to the tissues and cells, and the ECM of the skin is mainly composed of collagen, elastin and glycosaminoglycans (GAGs) 49 . Phosphatidylinositols are a class of phospholipids that have various roles in cytoskeleton organization 50 . Notably, the dysregulation of ECM structural constituent (crest 1: adjusted P = 3.32 × 10 −8 ; crest 2: adjusted P = 1.61 × 10 −8 ), GAG binding (crest 1: adjusted P = 1.805 × 10 −8 ; crest 2: adjusted P = 4.093 × 10 −6 ) and phosphatidylinositol binding (crest 1: adjusted P = 3.391 × 10 −6 ; crest 2: adjusted P = 7.832 × 10 −6 ) were identified 51 , 52 . We also identified cytolysis (crest 1: adjusted P = 2.973 × 10 −5 ; crest 2: adjusted P : NS), which can increase water loss 53 . The dysregulated actin binding (crest 1: adjusted P = 3.536 × 10 −8 ; crest 2: adjusted P = 3.435 × 10 −9 ), actin filament organization (crest 1: adjusted P = 8.406 × 10 −9 ; crest 2: adjusted P = 1.157 × 10 −9 ) and regulation of actin cytoskeleton (crest 1: adjusted P = 0.00090242; crest 2: adjusted P = 6.788 × 10 −4 ) were identified, which affect the structure and function of various tissues 54 , 55 , 56 , 57 , 58 . Additionally, cell adhesion is the attachment of a cell to another cell or to ECM via adhesion molecules 59 . We identified the positive regulation of cell adhesion (crest 1: adjusted P = 3.618 × 10 −5 ; crest 2: adjusted P = 8.272 × 10 −9 ) module, which can prevent or delay skin aging 60 , 61 . Threonine can affect sialic acid production, which is involved in cell adhesion 62 . We also identified the glycine, serine and threonine metabolism (crest 1: adjusted P : NS; crest 2: adjusted P = 0.00506) 62 . Additionally, scavenging of heme from plasma was identified (crest 1: adjusted P = 1.176 × 10 −11 ; crest 2: adjusted P = 1.694 × 10 −8 ), which can modulate skin aging as excess-free heme can damage cellular components 63 , 64 . Rho GTPases regulate a wide range of cellular responses, including changes to the cytoskeleton and cell adhesion (RHO GTPase cycle, crest 1: adjusted P = 9.956 × 10 −10 ; crest 2: adjusted P = 1.546 × 10 −5 ) 65 . In relation to muscle, previous studies demonstrated that muscle mass decreases by approximately 3–8% per decade after the age of 30, with an even higher decline rate after the age of 60, which consistently coincides with the observed second crest 66 . Interestingly, we identified dysregulation in the module associated with the structural constituent of muscle (crest 1: adjusted P = 0.00565; crest 2: adjusted P = 0.0162), consistent with previous findings 66 . Furthermore, we identified the pathway associated with caffeine metabolism (crest 1: adjusted P = 0.00378; crest 2: adjusted P = 0.0162), which is consistent with our observations above (Fig. 2b ) and implies that the capacity to metabolize caffeine undergoes a notable alteration not only around 60 years of age but also around the age of 40 years.
In crest 1, we identified specific modules associated with lipid and alcohol metabolism. Previous studies established that lipid metabolism declines with human aging 67 . Our analysis revealed several modules related to lipid metabolism, including plasma lipoprotein remodeling (crest 1: adjusted P = 2.269 × 10 −9 ), chylomicron assembly (crest 1: adjusted P = 9.065 × 10 −7 ) and ATP-binding cassette (ABC) transporters (adjusted P = 1.102 × 10 −4 ). Moreover, we discovered a module linked to alcohol metabolism (alcohol binding, adjusted P = 8.485 × 10 −7 ), suggesting a decline in alcohol metabolization efficiency with advancing age, particularly around the age of 40, when it significantly diminishes. In crest 2, we observed prominent modules related to immune dysfunction, encompassing acute-phase response (adjusted P = 2.851 × 10 −8 ), antimicrobial humoral response (adjusted P = 2.181 × 10 −5 ), zymogen activation (adjusted P = 4.367 × 10 −6 ), complement binding (adjusted P = 0.002568), mononuclear cell differentiation (adjusted P = 9.352 × 10 −8 ), viral process (adjusted P = 5.124 × 10 −7 ) and regulation of hemopoiesis (adjusted P = 3.522 × 10 −7 ) (Fig. 5a ). Age-related changes in the immune system, collectively known as immunosenescence, have been extensively documented 68 , 69 , 70 , and our results demonstrate a rapid decline at age 60. Furthermore, we also identified modules associated with kidney function (glomerular filtration, adjusted P = 0.00869) and carbohydrate metabolism (carbohydrate binding, adjusted P = 0.01045). Our previous findings indicated a decline in kidney function around the age of 60 years (Fig. 3c ), as did the present result of this observation. Previous studies indicated the influence of carbohydrates on aging, characterized by the progressive decline of physiological functions and increased susceptibility to diseases over time 71 , 72 .
In summary, our analysis identifies many dysregulated functional modules identified in both crest 1 and crest 2 that underlie the risk for various diseases and alterations of biological functions. Notably, we observed an overlap of dysregulated functional modules among clusters 2, 4 and 6 because they overlap at the molecular level, as depicted in Fig. 5b . This indicates that certain molecular components are shared among these clusters and the identified crests. However, it is important to note that numerous molecules are specific to each of the two approaches employed in our study. This suggests that these two approaches complement each other in identifying nonlinear changes in molecules and functions during human aging. By using both approaches, we were able to capture a more comprehensive understanding of the molecular alterations associated with aging and their potential implications for diseases.
Analyzing a longitudinal multi-omics dataset involving 108 participants, we successfully captured the dynamic and nonlinear molecular changes that occur during human aging. Our study’s strength lies in the comprehensive nature of the dataset, which includes multiple time point samples for each participant. This longitudinal design enhances the reliability and robustness of our findings compared to cross-sectional studies with only one time point sample for each participant. The first particularly intriguing finding from our analysis is that only a small fraction of molecules (6.6%) displayed linear changes throughout human aging (Fig. 1d ). This observation is consistent with previous research and underscores the limitations of relying solely on linear regression to understand the complexity of aging-related molecular changes 5 . Instead, our study revealed that a considerable number of molecules (81.0%) exhibited nonlinear patterns (Fig. 1e ). Notably, this nonlinear trend was observed across all types of omics data with remarkably high consistency (Fig. 1e,g ), highlighting the widespread functionally relevant nature of these dynamic changes. By unveiling the nonlinear molecular alterations associated with aging, our research contributes to a more comprehensive understanding of the aging process and its molecular underpinnings.
To further investigate the nonlinear changing molecules observed in our study, we employed a trajectory clustering approach to group molecules with similar temporal patterns. This analysis revealed the presence of three distinct clusters (Fig. 2c ) that exhibited clear and compelling patterns across the human lifespan. These clusters suggest that there are specific age ranges, such as around 60 years old, where distinct and extensive molecular changes occur (Fig. 2c ). Functional analysis revealed several modules that exhibited nonlinear changes during human aging. For example, we identified a module associated with oxidative stress, which is consistent with previous studies linking oxidative stress to the aging process 23 (Fig. 3a ). Our analysis indicates that this pathway increases significantly after the age of 60 years. In cluster 5, we identified a transcriptomics module associated with mRNA stabilization and autophagy (Fig. 3a ). Both of these processes have been implicated in the aging process and are involved in maintaining cellular homeostasis and removing damaged components. Furthermore, our analysis uncovered nonlinear changes in disease risk across aging. In cluster 2, we identified the phenylalanine metabolism pathway (Fig. 3b ), which has been associated with cardiac dysfunction during aging 26 . Additionally, we found that the clinical laboratory tests blood urea nitrogen and serum/plasma glucose increase significantly with age (cluster 2; Fig. 3c ), indicating a nonlinear decline in kidney function and an increased risk of T2D with age, with a critical threshold occurring approximately at the age of 60 years. In cluster 4, we identified pathways related to cardiovascular health, such as the biosynthesis of unsaturated fatty acids and caffeine metabolism (Fig. 3b ). Overall, our study provides compelling evidence for the existence of nonlinear changes in molecular profiles during human aging. By elucidating the specific functional modules and disease-related pathways that exhibit such nonlinear changes, we contribute to a better understanding of the complex molecular dynamics underlying the aging process and its implications for disease risk.
Although the trajectory clustering approach proves effective in identifying molecules that undergo nonlinear changes, it may not be as proficient in capturing substantial alterations that occur at specific time points without exhibiting a consistent pattern in other stages. We then employed a modified version of the DE-SWAN algorithm 14 to comprehensively investigate changes in multi-omics profiling throughout human aging. This approach enabled us to identify waves of dysregulated molecules and microbes across the human lifespan. Our analysis revealed two prominent crests occurring around the ages of 40 years and 60 years, which were consistent across various omics data types, suggesting their universal nature (Fig. 4a,e ). Notably, in the proteomics data, we observed crests around the ages of 40 years and 60 years, which aligns approximately with a previous study (which reported crests at ages 34 years, 60 years and 78 years) 14 . Due to the age range of our cohort being 25–75 years, we did not detect the third peak. Furthermore, the differences in proteomics data acquisition platforms (mass spectrometry versus SomaScan) 14 , 73 resulted in different identified proteins, with only a small overlap (1,305 proteins versus 302 proteins, of which only 75 were shared). This discrepancy may explain the age variation of the first crest identified in the two studies (approximately 10 years). However, despite the differences in the two proteomics datasets, the wave patterns observed in both studies were highly similar 14 (Fig. 4a ). Remarkably, by considering multiple omics data types, we consistently identified similar crests for each type, indicating the universality of these waves of change across plasma molecules and microbes from various body sites (Fig. 4e and Supplementary Fig. 3 ).
The analysis of molecular functionality in the two distinct crests revealed the presence of several modules, indicating a nonlinear increase in the risks of various diseases (Fig. 5a ). Both crest 1 and crest 2 exhibit the identification of multiple modules associated with CVD, which aligns with the aforementioned findings (Fig. 3b ). Moreover, we observed an escalated dysregulation in skin and muscle functioning in both crest 1 and crest 2. Additionally, we identified a pathway linked to caffeine metabolism, indicating a noticeable alteration in caffeine metabolization not only around the age of 60 but also around the age of 40. This shift may be due to either a metabolic shift or a change in caffeine consumption. In crest 1, we also identified specific modules associated with lipid and alcohol metabolism, whereas crest 2 demonstrated prominent modules related to immune dysfunction. Furthermore, we also detected modules associated with kidney function and carbohydrate metabolism, which is consistent with our above results. These findings reinforce our previous observations regarding a decline in kidney function around the age of 60 years (Fig. 3c ) while shedding light on the impact of dysregulated functional modules in both crest 1 and crest 2, suggesting nonlinear changes in disease risk and functional dysregulation. Notably, we identified an overlap of dysregulated functional modules among clusters 2, 4 and 6, indicating molecular-level similarities between these clusters and the identified crests (Fig. 5b ). This suggests the presence of shared molecular components among these clusters and crests. However, it is crucial to note that there are also numerous molecules specific to each of the two approaches employed in our study, indicating that these approaches complement each other in identifying nonlinear changes in molecules and functions during human aging.
The present research is subject to certain constraints. We accounted for many basic characteristics (confounders) of participants in the cohort; but because this study primarily reflects between-individual differences, there may be additional confounders due to the different age distributions of the participants. For example, we identified a notable decrease in oxygen carrier activity around age 60 (Figs. 2c and 3a ) and marked variations in alcohol and caffeine metabolism around ages 40 and 60 (Fig. 3a ). However, these findings might be shaped by participants’ lifestyle—that is, physical activity and their alcohol and caffeine intake. Regrettably, we do not have such detailed behavioral data for the entire group, necessitating validation in upcoming research. Although initial BMI and insulin sensitivity measurements were available at cohort entry, subsequent metrics during the observation span were absent, marking a study limitation.
A further constraint is our cohort’s modest size, encompassing merely 108 individuals (eight individuals between 25 years and 40 years of age), which hampers the full utilization of deep learning and may affect the robustness of the identification of nonlinear changing features in Fig. 1e . Although advanced computational techniques, including deep learning, are pivotal for probing nonlinear patterns, our sample size poses restrictions. Expanding the cohort size in subsequent research would be instrumental in harnessing the full potential of machine learning tools. Another limitation of our study is that the recruitment of participants was within the community around Stanford University, driven by rigorous sample collection procedures and the substantial expenses associated with setting up a longitudinal cohort. Although our participants exhibited a considerable degree of ethnic age and biological sex diversity (Fig. 1a and Supplementary Data ), it is important to acknowledge that our cohort may not fully represent the diversity of the broader population. The selectivity of our cohort limits the generalizability of our findings. Future studies should aim to include a more diverse cohort to enhance the external validity and applicability of the results.
In addition, the mean observation span for participants was 626 days, which is insufficient for detailed inflection point analyses. Our cohort’s age range of 25–70 years lacks individuals who lie outside of this range. The molecular nonlinearity detected might be subject to inherent variations or oscillations, a factor to consider during interpretation. Our analysis has not delved into the nuances of the dynamical systems theory, which provides a robust mathematical framework for understanding observed behaviors. Delving into this theory in future endeavors may yield enhanced clarity and interpretation of the data.
Moreover, it should be noted that, in our study, the observed nonlinear molecular changes occurred across individuals of varying ages rather than within the same individuals. This is attributed to the fact that, despite our longitudinal study, the follow-up period for our participants was relatively brief for following aging patterns (median, 1.7 years; Extended Data Fig. 1g ). Such a timeframe is inadequate for detecting nonlinear molecular changes that unfold over decades throughout the human lifespan. Addressing this limitation in future research is essential.
Lastly, our study’s molecular data are derived exclusively from blood samples, casting doubt on its direct relevance to specific tissues, such as the skin or muscles. We propose that blood gene expression variations might hint at overarching physiological alterations, potentially impacting the ECM in tissues, including skin and muscle. Notably, some blood-based biomarkers and transcripts have demonstrated correlations with tissue modifications, inflammation and other elements influencing the ECM across diverse tissues 74 , 75 .
In our future endeavors, the definitive confirmation of our findings hinges on determining if nonlinear molecular patterns align with nonlinear changes in functional capacities, disease occurrences and mortality hazards. For a holistic grasp of this, amalgamating multifaceted data from long-term cohort studies covering several decades becomes crucial. Such data should encompass molecular markers, comprehensive medical records, functional assessments and mortality data. Moreover, employing cutting-edge statistical techniques is vital to intricately decipher the ties between these nonlinear molecular paths and health-centric results.
In summary, the unique contribution of our study lies not merely in reaffirming the nonlinear nature of aging but also in the depth and breadth of the multi-omics data that we analyzed. Our study goes beyond stating that aging is nonlinear by identifying specific patterns, inflection points and potential waves in aging across multiple layers of biological data during human aging. Identifying specific clusters with distinct patterns, functional implications and disease risks enhances our understanding of the aging process. By considering the nonlinear dynamics of aging-related changes, we can gain insights into specific periods of significant changes (around age 40 and age 60) and the molecular mechanisms underlying age-related diseases, which could lead to the development of early diagnosis and prevention strategies. These comprehensive multi-omics data and the approach allow for a more nuanced understanding of the complexities involved in the aging process, which we think adds value to the existing body of research. However, further research is needed to validate and expand upon these findings, potentially incorporating larger cohorts to capture the full complexity of aging.
The participant recruitment, sample collection, data acquisition and data processing were documented in previous studies conducted by Zhou et al. 76 , Ahadi et al. 5 , Schüssler-Fiorenza Rose et al. 77 , Hornburg et al. 78 and Zhou et al. 79 .
Participant recruitment
Participants provided informed written consent for the study under research protocol 23602, which was approved by the Stanford University institutional review board. This study adheres to all relevant ethical regulations, ensuring informed consents were obtained from all participants. All participants consented to publication of potentially identifiable information. The cohort comprised 108 participants who underwent follow-up assessments. Exclusion criteria encompassed conditions such as anemia, kidney disease, a history of CVD, cancer, chronic inflammation or psychiatric illnesses as well as any prior bariatric surgery or liposuction. Each participant who met the eligibility criteria and provided informed consent underwent a one-time modified insulin suppression test to quantify insulin-mediated glucose uptake at the beginning of the enrollment 76 . The steady-state plasma glucose (SSPG) levels served as a direct indicator of each individual’s insulin sensitivity in processing a glucose load. We categorized individuals with SSPG levels below 150 mg dl −1 as insulin sensitive and those with levels of 150 mg dl −1 or higher as insulin resistant 80 , 81 . Thirty-eight participants were missing SSPG values, rendering their insulin resistance or sensitivity status undetermined. We also collected fasting plasma glucose (FPG) data for 69 participants at enrollment. Based on the FPG levels, two participants were identified as having diabetes at enrollment, with FPG levels exceeding 126 mg dl −1 ( Supplementary Data ). Additionally, we measured hemoglobin A1C (HbA1C) levels during each visit, using it as a marker for average glucose levels over the past 3 months: 6.5% or higher indicates diabetes. Accordingly, four participants developed diabetes during the study period. At the beginning of the enrollment, BMI was also measured for each participant. Participants received no compensation.
Comprehensive sample collection was conducted during the follow-up period, and multi-omics data were acquired (Fig. 1b ). For each visit, the participants self-reported as healthy or non-healthy 76 . To ensure accuracy and minimize the impact of confounding factors, only samples from individuals classified as healthy were selected for subsequent analysis.
Transcriptomics
Transcriptomic profiling was conducted on flash-frozen PBMCs. RNA isolation was performed using a QIAGEN All Prep kit. Subsequently, RNA libraries were assembled using an input of 500 ng of total RNA. In brief, ribosomal RNA (rRNA) was selectively eliminated from the total RNA pool, followed by purification and fragmentation. Reverse transcription was carried out using a random primer outfitted with an Illumina-specific adaptor to yield a cDNA library. A terminal tagging procedure was used to incorporate a second adaptor sequence. The final cDNA library underwent amplification. RNA sequencing libraries underwent sequencing on an Illumina HiSeq 2000 platform. Library quantification was performed via an Agilent Bioanalyzer and Qubit fluorometric quantification (Thermo Fisher Scientific) using a high-sensitivity dsDNA kit. After normalization, barcoded libraries were pooled at equimolar ratios into a multiplexed sequencing library. An average of 5–6 libraries were processed per HiSeq 2000 lane. Standard Illumina pipelines were employed for image analysis and base calling. Read alignment to the hg19 reference genome and personal exomes was achieved using the TopHat package, followed by transcript assembly and expression quantification via HTseq and DESeq2. In the realm of data pre-processing, genes with an average read count across all samples lower than 0.5 were excluded. Samples exhibiting an average read count lower than 0.5 across all remaining genes were likewise removed. For subsequent global variance and correlation assessments, genes with an average read count of less than 1 were eliminated.
Plasma sample tryptic peptides were fractionated using a NanoLC 425 System (SCIEX) operating at a flow rate of 5 μl min −1 under a trap-elute configuration with a 0.5 × 10 mm ChromXP column (SCIEX). The liquid chromatography gradient was programmed for a 43-min run, transitioning from 4% to 32% of mobile phase B, with an overall run time of 1 h. Mobile phase A consisted of water with 0.1% formic acid, and mobile phase B was formulated with 100% acetonitrile and 0.1% formic acid. An 8-μg aliquot of non-depleted plasma was loaded onto a 15-cm ChromXP column. Mass spectrometry analysis was executed employing SWATH acquisition on a TripleTOF 6600 system. A set of 100 variable Q1 window SWATH acquisition methods was designed in high-sensitivity tandem mass spectrometry (MS/MS) mode. Subsequent data analysis included statistical scoring of peak groups from individual runs via pyProphet 82 , followed by multi-run alignment through TRIC60, ultimately generating a finalized data matrix with a false discovery rate (FDR) of 1% at the peptide level and 10% at the protein level. Protein quantitation was based on the sum of the three most abundant peptide signals for each protein. Batch effect normalization was achieved by subtracting principal components that primarily exhibited batch-associated variation, using Perseus software v.1.4.2.40.
Untargeted metabolomics
A ternary solvent system of acetone, acetonitrile and methanol in a 1:1:1 ratio was used for metabolite extraction. The extracted metabolites were dried under a nitrogen atmosphere and reconstituted in a 1:1 methanol:water mixture before analysis. Metabolite profiles were generated using both hydrophilic interaction chromatography (HILIC) and reverse-phase liquid chromatography (RPLC) under positive and negative ion modes. Thermo Q Exactive Plus mass spectrometers were employed for HILIC and RPLC analyses, respectively, in full MS scan mode. MS/MS data were acquired using quality control (QC) samples. For the HILIC separations, a ZIC-HILIC column was used with mobile phase solutions of 10 mM ammonium acetate in 50:50 and 95:5 acetonitrile:water ratios. In the case of RPLC, a Zorbax SBaq column was used, and the mobile phase consisted of 0.06% acetic acid in water and methanol. Metabolic feature detection was performed using Progenesis QI software. Features from blanks and those lacking sufficient linearity upon dilution were excluded. Only features appearing in more than 33% of the samples were retained for subsequent analyses, and any missing values were imputed using the k -nearest neighbors approach. We employed locally estimated scatterplot smoothing (LOESS) normalization 83 to correct the metabolite-specific signal drift over time. The metid package 84 was used for metabolite annotation.
Cytokine data
A panel of 62 human cytokines, chemokines and growth factors was analyzed in EDTA-anticoagulated plasma samples using Luminex-based multiplex assays with conjugated antibodies (Affymetrix). Raw fluorescence measurements were standardized to median fluorescence intensity values and subsequently subjected to variance-stabilizing transformation to account for batch-related variations. As previously reported 76 , data points characterized by background noise, termed CHEX, that deviate beyond five standard deviations from the mean (mean ± 5 × s.d.) were excluded from the analyses.
Clinical laboratory test
The tests encompassed a comprehensive metabolic panel, a full blood count, glucose and HbA1C levels, insulin assays, high-sensitivity C-reactive protein (hsCRP), immunoglobulin M (IgM) and lipid, kidney and liver panels.
Lipid extraction and quantification procedures were executed in accordance with established protocols 78 . In summary, complex lipids were isolated from 40 μl of EDTA plasma using a solvent mixture comprising methyl tertiary-butyl ether, methanol and water, followed by a biphasic separation. Subsequent lipid analysis was conducted on the Lipidyzer platform, incorporating a differential mobility spectrometry device (SelexION Technology) and a QTRAP 5500 mass spectrometer (SCIEX).
Immediately after arrival, samples were stored at −80 °C. Stool and nasal samples were processed and sequenced in-house at the Jackson Laboratory for Genomic Medicine, whereas oral and skin samples were outsourced to uBiome for additional processing. Skin and oral samples underwent 30 min of beads-beating lysis, followed by a silica-guanidinium thiocyanate-based nucleic acid isolation protocol. The V4 region of the 16S rRNA gene was amplified using specific primers, after which the DNA was barcoded and sequenced on an Illumina NextSeq 500 platform via a 2 × 150-bp paired-end protocol. Similarly, stool and nasal samples were processed for 16S rRNA V1–V3 region amplification using a different set of primers and sequenced on an Illumina MiSeq platform. For data processing, the raw sequencing data were demultiplexed using BCL2FASTQ software and subsequently filtered for quality. Reads with a Q-score lower than 30 were excluded. The DADA2 R package was used for further sequence data processing, which included filtering out reads with ambiguous bases and errors, removing chimeras and aligning sequences against a validated 16S rRNA gene database. Relative abundance calculations for amplicon sequence variants (ASVs) were performed, and samples with inadequate sequencing depth (<1,000 reads) were excluded. Local outlier factor (LOF) was calculated for each point on a depth-richness plot, and samples with abnormal LOF were removed. In summary, rigorous procedures were followed in both the collection and processing stages, leveraging automated systems and specialized software to ensure the quality and integrity of the microbiome data across multiple body sites.
Statistics and reproducibility
For all data processing, statistical analysis and data visualization tasks, RStudio, along with R language (v.4.2.1), was employed. A comprehensive list of the packages used can be found in the Supplementary Note . The Benjamini–Hochberg method was employed to account for multiple comparisons. Spearman correlation coefficients were calculated using the R functions ‘cor’ and ‘cor.test’. Principal-component analysis (PCA) was conducted using the R function ‘princomp’. Before all the analyses, the confounders, such as BMI, sex, IRIS and ethnicity, were adjusted using the previously published method 19 . In brief, we used the intensity of each feature as the dependent variable (Y) and the confounding factors as the independent variables (X) to build a linear regression model. The residuals from this model were then used as the adjusted values for that specific feature.
All the omics data were acquired randomly. No statistical methods were used to predetermine the sample size, but our sample sizes are similar to those reported in previous publications 5 , 76 , 77 , 78 , 79 , and no data were excluded from the analyses. Additionally, the investigators were blinded to allocation during experiments and outcome assessment to the conditions of the experiments. Data distribution was assumed to be normal, but this was not formally tested.
The icons used in figures are from iconfont.cn, which can be used for non-commercial purposes under the MIT license ( https://pub.dev/packages/iconfont/license ).
Cross-sectional dataset generation
The ‘cross-sectional’ dataset was created by briefly extracting information from the longitudinal dataset. The mean value was calculated to represent each molecule’s intensity for each participant. Similarly, the age of each participant was determined by calculating the mean value of ages across all sample collection time points.
Linear changing molecule detection
We detected linear changing molecules during human aging using Spearman correlation and linear regression modeling. The confounders, such as BMI, sex, IRIS and ethnicity, were adjusted using the previously published method 19 . Our analysis revealed a high correlation between these two approaches in identifying such molecules. Based on these findings, we used the Spearman correlation approach to showcase the linear changing molecules during human aging. The permutation test was also used to get the permutated P values for each feature. In brief, each feature was subjected to sample label shuffling followed by a recalculation of the Spearman correlation. This process was reiterated 10,000 times, yielding 10,000 permuted Spearman correlations. The original Spearman correlation was then compared against these permuted values to obtain the permuted P values.
Dysregulated molecules compared to baseline during human aging
To depict the dysregulated molecules during human aging compared to the baseline, we categorized the participants into different age stages based on their ages. The baseline stage was defined as individuals aged 25–40 years. For each age stage group, we employed the Wilcoxon test to identify dysregulated molecules in comparison to the baseline, considering a significance threshold of P < 0.05. Before the statistical analysis, all the confounders were corrected. Subsequently, we visualized the resulting dysregulated molecules at different age stages using a Sankey plot. The permutation test was also used to get the permutated P values for each feature. In brief, we shuffled the sample labels and recalculated the absolute mean difference between the two groups, against which the actual absolute mean difference was benchmarked to derive the permuted P values. To identify the molecules and microbes that exhibited significant changes at any given age stage, we adjusted the P values for each feature by multiplying them by 6. This adjustment adheres to the Bonferroni correction method, ensuring a rigorous evaluation of statistical significance.
Evaluation of the age reflected by different types of omics data
To assess whether each type of omics data accurately reflects the ages of individuals in our dataset, we conducted a PCA. Subsequently, we computed the Spearman correlation coefficient between the ages of participants and the first principal component (PC1). The absolute value of this coefficient was used to evaluate the degree to which the omics data reflect the ages (Fig. 2a ). PLS regression was also used to compare the strength of the age effect to the different omics data types. In brief, the ‘pls’ function from the R package mixOmics was used to construct the regression model between omics data and ages. Then, the ‘perf’ function was used to assess the performance of all the modules with sevenfold cross-validation. The R 2 was extracted to assess the strength of the age effect on the different omics data types.
To accommodate the varying time points of biological and omics data, we employed the LOESS approach. This approach allowed us to smooth and predict the multi-omics data at specific time points (that is, every half year) 14 , 85 . In brief, for each molecule, we fitted a LOESS regression model. During the fitting process, the LOESS argument ‘span’ was optimized through cross-validation. This ensured that the LOESS model provided an accurate and non-overfitting fit to the data (Supplementary Fig. 2a,b ). Once we obtained the LOESS prediction model, we applied it to predict the intensity of each molecule at every half-year time point.
Trajectory clustering analysis
To conduct trajectory clustering analysis, we employed the fuzzy c-means clustering approach available in the R package ‘Mfuzz.’ This approach was previously described in our publication 19 . The analysis proceeded in several steps. First, the omics data were auto-scaled to ensure comparable ranges. Next, we computed the minimum centroid distances for a range of cluster numbers, specifically from 2 to 22, in step 1. These minimum centroid distances served as a cluster validity index, helping us determine the optimal cluster number. Based on predefined rules, we selected the optimal cluster number. To refine the accuracy of this selection, we merged clusters with center expression data correlations greater than 0.8 into a single cluster. This step aimed to capture similar patterns within the data. The resulting optimal cluster number was then used for the fuzzy c-means clustering. Only molecules with memberships above 0.5 were retained within each cluster for further analysis. This threshold ensured that the molecules exhibited a strong association with their assigned cluster and contributed considerably to the cluster’s characteristics.
Pathway enrichment analysis and functional module identification
Transcriptomics and proteomics pathway enrichment.
Pathway enrichment analysis was conducted using the ‘clusterProfiler’ R package 86 . The GO, KEGG and Reactome databases were used. The P values were adjusted using the Benjamini–Hochberg method, with a significance threshold set at <0.05. To minimize redundant enriched pathways and GO terms, we employed a series of analyses. First, for enriched GO terms, we used the ‘Wang’ algorithm from the R package ‘simplifyEnrichment’ to calculate the similarity between GO terms. Only connections with a similarity score greater than 0.7 were retained to construct the GO term similarity network. Subsequently, community analysis was performed using the ‘igraph’ R package to partition the network into distinct modules. The GO term with the smallest enrichment adjusted P value was chosen as the representative within each module. The same approach was applied to the enriched KEGG and Reactome pathways, with one slight modification. In this case, the ‘jaccard’ algorithm was used to calculate the similarity between pathways, and a similarity cutoff of 0.5 was employed for the Jaccard index. After removing redundant enriched pathways, we combined all the remaining GO terms and pathways. Subsequently, we calculated the similarity between these merged entities using the Jaccard index. This similarity analysis aimed to capture the overlap and relationships between the different GO terms and pathways. Using the same approach as before, we performed community analysis to identify distinct biological functional modules based on the merged GO terms and pathways.
Identification of functional modules
First, we used the ‘Wang’ algorithm for the GO database and the ‘jaccard’ algorithm for the KEGG and Reactome databases to calculate the similarity between pathways. The enriched pathways served as nodes in a similarity network, with edges representing the similarity between two nodes. Next, we employed the R package ‘igraph’ to identify modules within the network based on edge betweenness. By gradually removing edges with the highest edge betweenness scores, we constructed a hierarchical map known as a dendrogram, representing a rooted tree of the graph. The leaf nodes correspond to individual pathways, and the root node represents the entire graph 87 . We then merged pathways within each module, selecting the pathway with the smallest adjusted P value to represent the module. After this step, we merged pathways from all three databases into modules. Subsequently, we repeated the process by calculating the similarity between modules from all three databases using the ‘jaccard’ algorithm. Once again, we employed the same approach described above to identify the functional modules.
Metabolomics pathway enrichment
To perform pathway enrichment analysis for metabolomics data, we used the human KEGG pathway database. This database was obtained from KEGG using the R package ‘massDatabase’ 88 . For pathway enrichment analysis, we employed the hypergeometric distribution test from the ‘TidyMass’ project 89 . This statistical test allowed us to assess the enrichment of metabolites within each pathway. To account for multiple tests, P values were adjusted using the Benjamini–Hochberg method. We considered pathways with Benjamini–Hochberg-adjusted P values lower than 0.05 as significantly enriched.
Modified DE-SWAN
The DE-SWAN algorithm 14 was used. To begin, a unique age is selected as the center of a 20-year window. Molecule levels in individuals younger than and older than that age are compared using the Wilcoxon test to assess differential expression. P values are calculated for each molecule, indicating the significance of the observed differences. To ensure sufficient sample sizes for statistical analysis in each time window, the initial window ranges from ages 25 to 50. The left half of this window covers ages 25–40, whereas the right half spans ages 41–50. The window then moves in one-year steps; this is why Fig. 4 displays an age range of 40–65 years. To account for multiple comparisons, these P values are adjusted using Benjamini–Hochberg correction. To evaluate the robustness and relevance of the DE-SWAN results, the algorithm is tested with various parcel widths, including 15 years, 20 years, 25 years and 30 years. Additionally, different q value thresholds, such as <0.0001, <0.001, <0.01 and <0.05, are applied. By comparing the results obtained with these different parameters to results obtained by chance, we can assess the significance of the findings. To generate random results for comparison, the phenotypes of the individuals are randomly permuted, and the modified DE-SWAN algorithm is applied to the permuted dataset. This allows us to determine whether the observed results obtained with DE-SWAN are statistically significant and not merely a result of chance.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw data used in this study can be accessed without any restrictions on the National Institutes of Health Human Microbiome 2 project site ( https://portal.hmpdacc.org ). Both the raw and processed data are also available on the Stanford iPOP site ( http://med.stanford.edu/ipop.html ). Researchers and interested individuals can visit these websites to access the data. For further details and inquiries about the study, we recommend contacting the corresponding author, who can provide additional information and address any specific questions related to the research.
Code availability
The statistical analysis and data processing in this study were performed using R v.4.2.1, along with various base packages and additional packages. Detailed information about the specific packages used can be found in the Supplementary Note , which accompanies the manuscript. Furthermore, all the custom scripts developed for this study have been made openly accessible and can be found on the GitHub repository at https://github.com/jaspershen-lab/ipop_aging . By visiting this repository, researchers and interested individuals can access and use the custom scripts for their own analyses or to replicate the study’s findings.
Hou, Y. et al. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 15 , 565–581 (2019).
Article PubMed Google Scholar
Chen, R. et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148 , 1293–1307 (2012).
Article CAS PubMed PubMed Central Google Scholar
Valdes, A. M., Glass, D. & Spector, T. D. Omics technologies and the study of human ageing. Nat. Rev. Genet. 14 , 601–607 (2013).
Rutledge, J., Oh, H. & Wyss-Coray, T. Measuring biological age using omics data. Nat. Rev. Genet. 23 , 715–727 (2022).
Ahadi, S. et al. Personal aging markers and ageotypes revealed by deep longitudinal profiling. Nat. Med. 26 , 83–90 (2020).
Ram, U. et al. Age-specific and sex-specific adult mortality risk in India in 2014: analysis of 0.27 million nationally surveyed deaths and demographic estimates from 597 districts. Lancet Glob. Health 3 , e767–e775 (2015).
Rodgers, J. L. et al. Cardiovascular risks associated with gender and aging. J. Cardiovasc. Dev. Dis. 6 , 19 (2019).
CAS PubMed PubMed Central Google Scholar
Poewe, W. et al. Parkinson disease. Nat. Rev. Dis. Primers 3 , 17013 (2017).
Hy, L. X. & Keller, D. M. Prevalence of AD among whites: a summary by levels of severity. Neurology 55 , 198–204 (2000).
Article CAS PubMed Google Scholar
Nussbaum, R. L. & Ellis, C. E. Alzheimer’s disease and Parkinson’s disease. N. Engl. J. Med. 348 , 1356–1364 (2003).
Xiong, Y. et al. Vimar/RAP1GDS1 promotes acceleration of brain aging after flies and mice reach middle age. Commun. Biol. 6 , 420 (2023).
Sherwood, C. C. et al. Aging of the cerebral cortex differs between humans and chimpanzees. Proc. Natl Acad. Sci. USA 108 , 13029–13034 (2011).
Márquez, E. J. et al. Sexual-dimorphism in human immune system aging. Nat. Commun. 11 , 751 (2020).
Article PubMed PubMed Central Google Scholar
Lehallier, B. et al. Undulating changes in human plasma proteome profiles across the lifespan. Nat. Med. 25 , 1843–1850 (2019).
Fehlmann, T. et al. Common diseases alter the physiological age-related blood microRNA profile. Nat. Commun. 11 , 5958 (2020).
Shavlakadze, T. et al. Age-related gene expression signature in rats demonstrate early, late, and linear transcriptional changes from multiple tissues. Cell Rep. 28 , 3263–3273 (2019).
Vershinina, O., Bacalini, M. G., Zaikin, A., Franceschi, C. & Ivanchenko, M. Disentangling age-dependent DNA methylation: deterministic, stochastic, and nonlinear. Sci Rep. 11 , 9201 (2021).
Li, J. et al. Determining a multimodal aging clock in a cohort of Chinese women. Med 4 , 825–848 (2023).
Shen, X. Multi-omics microsampling for the profiling of lifestyle-associated changes in health. Nat. Biomed. Eng . 8 , 11–29 (2024).
Takahashi, T. A. & Johnson, K. M. Menopause. Med. Clin. North Am. 99 , 521–534 (2015).
Umbayev, B. et al. Role of a small GTPase Cdc42 in aging and age-related diseases. Biogerontology 24 , 27–46 (2023).
Yi, S.-J. & Kim, K. New insights into the role of histone changes in aging. Int. J. Mol. Sci. 21 , 8241 (2020).
Liguori, I. et al. Oxidative stress, aging, and diseases. Clin. Interv. Aging 13 , 757–772 (2018).
Borbolis, F. & Syntichaki, P. Cytoplasmic mRNA turnover and ageing. Mech. Ageing Dev. 152 , 32–42 (2015).
Kaushik, S. et al. Autophagy and the hallmarks of aging. Ageing Res. Rev. 72 , 101468 (2021).
Czibik, G. et al. Dysregulated phenylalanine catabolism plays a key role in the trajectory of cardiac aging. Circulation 144 , 559–574 (2021).
Rousselle, A. et al. CXCL5 limits macrophage foam cell formation in atherosclerosis. J. Clin. Invest. 123 , 1343–1347 (2013).
Fazeli, P. K., Lee, H. & Steinhauser, M. L. Aging is a powerful risk factor for type 2 diabetes mellitus independent of body mass index. Gerontology 66 , 209–210 (2019).
Allayee, H., Roth, N. & Hodis, H. N. Polyunsaturated fatty acids and cardiovascular disease: implications for nutrigenetics. J. Nutrigenet. Nutrigenomics 2 , 140–148 (2009).
Sacks, F. M. et al. Dietary fats and cardiovascular disease: a presidential advisory from the American Heart Association. Circulation 136 , e1–e23 (2017).
Qi, W. et al. The ω-3 fatty acid α-linolenic acid extends Caenorhabditis elegans lifespan via NHR-49/PPARα and oxidation to oxylipins. Aging Cell 16 , 1125–1135 (2017).
Bird, A. W. et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419 , 411–415 (2002).
Sivanand, S. et al. Nuclear acetyl-CoA production by ACLY promotes homologous recombination. Mol. Cell 67 , 252–265 (2017).
Zhao, S. et al. Regulation of cellular metabolism by protein lysine acetylation. Science 327 , 1000–1004 (2010).
Vericel, E. et al. Platelets and aging I.—Aggregation, arachidonate metabolism and antioxidant status. Thromb. Res. 49 , 331–342 (1988).
Gu, S. X. & Dayal, S. Redox mechanisms of platelet activation in aging. Antioxidants (Basel) 11 , 995 (2022).
Oikonomopoulou, K., Ricklin, D., Ward, P. A. & Lambris, J. D. Interactions between coagulation and complement–their role in inflammation. Semin. Immunopathol. 34 , 151–165 (2012).
Wasiak, S. et al. Downregulation of the complement cascade in vitro, in mice and in patients with cardiovascular disease by the bet protein inhibitor apabetalone (RVX-208). J. Cardiovasc. Transl. 10 , 337–347 (2017).
Article Google Scholar
Slack, M. A. & Gordon, S. M. Protease activity in vascular disease. Arterioscl. Thromb. Vasc. Biol. 39 , e210–e218 (2019).
Mari, D. et al. Hemostasis and ageing. Immun. Ageing 5 , 12 (2008).
Lowe, G. & Rumley, A. The relevance of coagulation in cardiovascular disease: what do the biomarkers tell us? Thromb. Haemostasis 112 , 860–867 (2014).
Li, Y. et al. Branched chain amino acids exacerbate myocardial ischemia/reperfusion vulnerability via enhancing GCN2/ATF6/PPAR-α pathway-dependent fatty acid oxidation. Theranostics 10 , 5623–5640 (2020).
McGarrah, R. W. & White, P. J. Branched-chain amino acids in cardiovascular disease. Nat. Rev. Cardiol. 20 , 77–89 (2023).
Arsenian, M. Potential cardiovascular applications of glutamate, aspartate, and other amino acids. Clin. Cardiol. 21 , 620–624 (1998).
Grajeda-Iglesias, C. & Aviram, M. Specific amino acids affect cardiovascular diseases and atherogenesis via protection against macrophage foam cell formation: review article. Rambam Maimonides Med. J. 9 , e0022 (2018).
Chen, H. et al. Comprehensive metabolomics identified the prominent role of glycerophospholipid metabolism in coronary artery disease progression. Front. Mol. Biosci. 8 , 632950 (2021).
Giammanco, A. et al. Hyperalphalipoproteinemia and beyond: the role of HDL in cardiovascular diseases. Life (Basel) 11 , 581 (2021).
CAS PubMed Google Scholar
Zhu, Q. et al. Comprehensive metabolic profiling of inflammation indicated key roles of glycerophospholipid and arginine metabolism in coronary artery disease. Front. Immunol. 13 , 829425 (2022).
Yue, B. Biology of the extracellular matrix: an overview. J. Glaucoma 23 , S20–S23 (2014).
Zambrzycka, A. Aging decreases phosphatidylinositol-4,5-bisphosphate level but has no effect on activities of phosphoinositide kinases. Pol. J. Pharmacol. 56 , 651–654 (2004).
Lee, D. H., Oh, J.-H. & Chung, J. H. Glycosaminoglycan and proteoglycan in skin aging. J. Dermatol. Sci. 83 , 174–181 (2016).
Khan, A. U., Qu, R., Fan, T., Ouyang, J. & Dai, J. A glance on the role of actin in osteogenic and adipogenic differentiation of mesenchymal stem cells. Stem Cell Res. Ther. 11 , 283 (2020).
Lago, J. C. & Puzzi, M. B. The effect of aging in primary human dermal fibroblasts. PLoS ONE 14 , e0219165 (2019).
Pollard, T. D. Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 8 , a018226 (2016).
Lai, W.-F. & Wong, W.-T. Roles of the actin cytoskeleton in aging and age-associated diseases. Ageing Res. Rev. 58 , 101021 (2020).
Garcia, G., Homentcovschi, S., Kelet, N. & Higuchi-Sanabria, R. Imaging of actin cytoskeletal integrity during aging in C. elegans . Methods Mol. Biol. 2364 , 101–137 (2022).
Kim, Y. J. et al. Links of cytoskeletal integrity with disease and aging. Cells 11 , 2896 (2022).
Oosterheert, W., Klink, B. U., Belyy, A., Pospich, S. & Raunser, S. Structural basis of actin filament assembly and aging. Nature 611 , 374–379 (2022).
Bruzzone, A. et al. Dosage-dependent regulation of cell proliferation and adhesion through dual β2-adrenergic receptor/cAMP signals. FASEB J. 28 , 1342–1354 (2014).
McEver, R. P. & Luscinskas, F. W. Cell adhesion. In Hematology 7th edn (eds Hoffman, R. et al.) 127–134 (Elsevier, 2018).
Persa, O. D., Koester, J. & Niessen, C. M. Regulation of cell polarity and tissue architecture in epidermal aging and cancer. J. Invest. Dermatol. 141 , 1017–1023 (2021).
Canfield, C.-A. & Bradshaw, P. C. Amino acids in the regulation of aging and aging-related diseases. Transl. Med. Aging 3 , 70–89 (2019).
Chiabrando, D., Vinchi, F., Fiorito, V., Mercurio, S. & Tolosano, E. Heme in pathophysiology: a matter of scavenging, metabolism and trafficking across cell membranes. Front. Pharmacol. 5 , 61 (2014).
Aggarwal, S. et al. Heme scavenging reduces pulmonary endoplasmic reticulum stress, fibrosis, and emphysema. JCI Insight 3 , e120694 (2018).
Hodge, R. G. & Ridley, A. J. Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 17 , 496–510 (2016).
Siparsky, P. N., Kirkendall, D. T. & Garrett, W. E. Muscle changes in aging. Sports Health 6 , 36–40 (2014).
Johnson, A. A. & Stolzing, A. The role of lipid metabolism in aging, lifespan regulation, and age-related disease. Aging Cell 18 , e13048 (2019).
Paganelli, R., Scala, E., Quinti, I. & Ansotegui, I. J. Humoral immunity in aging. Aging Clin. Exp. Res. 6 , 143–150 (1994).
Article CAS Google Scholar
Goronzy, J. J. & Weyand, C. M. Understanding immunosenescence to improve responses to vaccines. Nat. Immunol. 14 , 428–436 (2013).
Cunha, L. L., Perazzio, S. F., Azzi, J., Cravedi, P. & Riella, L. V. Remodeling of the immune response with aging: immunosenescence and its potential impact on COVID-19 immune response. Front. Immunol. 11 , 1748 (2020).
Lee, D., Son, H. G., Jung, Y. & Lee, S.-J. V. The role of dietary carbohydrates in organismal aging. Cell. Mol. Life Sci. 74 , 1793–1803 (2017).
Franco-Juárez, B. et al. Effects of high dietary carbohydrate and lipid intake on the lifespan of C. elegans . Cells 10 , 2359 (2021).
Gold, L. et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 5 , e15004 (2010).
Galliera, E., Tacchini, L. & Corsi Romanelli, M. M. Matrix metalloproteinases as biomarkers of disease: updates and new insights. Clin. Chem. Lab. Med. 53 , 349–355 (2015).
Golusda, L., Kühl, A. A., Siegmund, B. & Paclik, D. Extracellular matrix components as diagnostic tools in inflammatory bowel disease. Biology (Basel) 10 , 1024 (2021).
Zhou, W. et al. Longitudinal multi-omics of host–microbe dynamics in prediabetes. Nature 569 , 663–671 (2019).
Schüssler-Fiorenza Rose, et al. A longitudinal big data approach for precision health. Nat. Med. 25 , 792–804 (2019).
Hornburg, D. et al. Dynamic lipidome alterations associated with human health, disease and ageing. Nat. Metab. 5 , 1578–1594 (2023).
Zhou, X. et al. Longitudinal profiling of the microbiome at four body sites reveals core stability and individualized dynamics during health and disease. Cell Host Microbe 32 , 506–526 (2024).
Contreras, P. H., Serrano, F. G., Salgado, A. M. & Vigil, P. Insulin sensitivity and testicular function in a cohort of adult males suspected of being insulin-resistant. Front. Med. (Lausanne) 5 , 190 (2018).
Evans, D. J., Murray, R. & Kissebah, A. H. Relationship between skeletal muscle insulin resistance, insulin-mediated glucose disposal, and insulin binding. Effects of obesity and body fat topography. J. Clin. Invest. 74 , 1515–1525 (1984).
Röst, H. L., Schmitt, U., Aebersold, R. & Malmström, L. pyOpenMS: a Python-based interface to the OpenMS mass-spectrometry algorithm library. Proteomics 14 , 74–77 (2014).
Shen, X. et al. Normalization and integration of large-scale metabolomics data using support vector regression. Metabolomics 12 , 89 (2016).
Shen, X. et al. metID: an R package for automatable compound annotation for LC−MS-based data. Bioinformatics 38 , 568–569 (2022).
Marabita, F. et al. Multiomics and digital monitoring during lifestyle changes reveal independent dimensions of human biology and health. Cell Syst. 13 , 241–255 (2022).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb.) 2 , 100141 (2021).
Newman, M. E. J. & Girvan, M. Finding and evaluating community structure in networks. Phys. Rev. E 69 , 026113 (2004).
Shen, X., Wang, C. & Snyder, M. P. massDatabase: utilities for the operation of the public compound and pathway database. Bioinformatics 38 , 4650–4651 (2022).
Shen, X. et al. TidyMass an object-oriented reproducible analysis framework for LC–MS data. Nat. Commun. 13 , 4365 (2022).
Download references
Acknowledgements
We sincerely thank all the research participants for their dedicated involvement in this study. We also thank A. Chen and L. Stainton for their valuable administrative assistance. Additionally, we are deeply grateful to A.T. Brunger’s support for this work. This work was supported by National Institutes of Health (NIH) grants U54DK102556 (M.P.S.), R01 DK110186-03 (M.P.S.), R01HG008164 (M.P.S.), NIH S10OD020141 (M.P.S.), UL1 TR001085 (M.P.S.) and P30DK116074 (M.P.S.) and by the Stanford Data Science Initiative (M.P.S.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
These authors contributed equally: Xiaotao Shen, Chuchu Wang.
Authors and Affiliations
Department of Genetics, Stanford University School of Medicine, Stanford, CA, USA
Xiaotao Shen, Xin Zhou, Wenyu Zhou, Daniel Hornburg, Si Wu & Michael P. Snyder
Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Singapore
Xiaotao Shen
School of Chemistry, Chemical Engineering and Biotechnology, Singapore, Singapore
Howard Hughes Medical Institute, Stanford University, Stanford, CA, USA
Chuchu Wang
Department of Molecular and Cellular Physiology, Stanford University, Stanford, CA, USA
Stanford Center for Genomics and Personalized Medicine, Stanford, CA, USA
Xin Zhou & Michael P. Snyder
You can also search for this author in PubMed Google Scholar
Contributions
X.S. and M.P.S. conceptualized and designed the study. X.Z. and W.Z. prepared the microbiome data. D.H. and W.S. prepared the lipidomics data. X.S. and C.W. conducted the data analysis. X.S. and C.W. prepared the figures. X.S., C.W. and M.P.S. contributed to the writing and revision of the manuscript, with input from other authors. M.S. and X.S. supervised the overall study.
Corresponding author
Correspondence to Michael P. Snyder .
Ethics declarations
Competing interests.
M.P.S. is a co-founder of Personalis, SensOmics, Qbio, January AI, Filtricine, Protos and NiMo and is on the scientific advisory boards of Personalis, SensOmics, Qbio, January AI, Filtricine, Protos, NiMo and Genapsys. D.H. has a financial interest in PrognomIQ and Seer. All other authors have no competing interests.
Peer review
Peer review information.
Nature Aging thanks Daniel Belsky and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended data fig. 1 demographic data of all the participants in the study..
a , The ages positively correlate with BMI. The shaded area around the regression line represents the 95% confidence interval. b , Gender with age. c , Ethnicity with age. d , Insulin response with age. e , biological sample collection for all the participants. f , Overlap of the different kinds of omics data. g , The age range for each participant in this study.
Extended Data Fig. 2 Most of the molecules change nonlinearly during human aging.
a , Differential expressional microbes in different age ranges compared to baselines (25 – 40 years old, two-sided Wilcoxon test, p -value < 0.05). b , Most of the linear changing molecules and microbiota are also included in the molecules/microbes that significantly dysregulated at least one age range.
Extended Data Fig. 3 Omics data can represent aging.
PCA score plot of metabolomics data ( a ), cytokine ( b ), and oral microbiome ( c ).
Extended Data Fig. 4 Functional analysis of molecules in different clusters.
a , The Jaccard index between clusters from different datasets. b , The overlap between clusters using different types of omics data. c , Functional module detection and identification. d , Functional analysis of nonlinear changing molecules for all clusters.
Extended Data Fig. 5 Function annotation for significantly dysregulated molecules in crest 1 and 2.
a , Transcriptomics data. b , Proteomics data. c , Metabolomics data.
Extended Data Fig. 6 Pathways enrichment results for crest 1 and 2.
a , The final functional modules identified for Crest 1 and 2. b , The pathway enrichment analysis results for transcriptomics data. c , The pathway enrichment analysis results for proteomics data. d , The pathway enrichment results for metabolomics data.
Supplementary information
Supplementary figs. 1–6, reporting summary, supplementary data analysis results of the study., rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Shen, X., Wang, C., Zhou, X. et al. Nonlinear dynamics of multi-omics profiles during human aging. Nat Aging (2024). https://doi.org/10.1038/s43587-024-00692-2
Download citation
Received : 09 December 2023
Accepted : 22 July 2024
Published : 14 August 2024
DOI : https://doi.org/10.1038/s43587-024-00692-2
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
- See us on facebook
- See us on twitter
- See us on youtube
- See us on linkedin
- See us on instagram
Massive biomolecular shifts occur in our 40s and 60s, Stanford Medicine researchers find
Time marches on predictably, but biological aging is anything but constant, according to a new Stanford Medicine study.
August 14, 2024 - By Rachel Tompa

We undergo two periods of rapid change, averaging around age 44 and age 60, according to a Stanford Medicine study. Ratana21 /Shutterstock.com
If it’s ever felt like everything in your body is breaking down at once, that might not be your imagination. A new Stanford Medicine study shows that many of our molecules and microorganisms dramatically rise or fall in number during our 40s and 60s.
Researchers assessed many thousands of different molecules in people from age 25 to 75, as well as their microbiomes — the bacteria, viruses and fungi that live inside us and on our skin — and found that the abundance of most molecules and microbes do not shift in a gradual, chronological fashion. Rather, we undergo two periods of rapid change during our life span, averaging around age 44 and age 60. A paper describing these findings was published in the journal Nature Aging Aug. 14.
“We’re not just changing gradually over time; there are some really dramatic changes,” said Michael Snyder , PhD, professor of genetics and the study’s senior author. “It turns out the mid-40s is a time of dramatic change, as is the early 60s. And that’s true no matter what class of molecules you look at.”
Xiaotao Shen, PhD, a former Stanford Medicine postdoctoral scholar, was the first author of the study. Shen is now an assistant professor at Nanyang Technological University Singapore.
These big changes likely impact our health — the number of molecules related to cardiovascular disease showed significant changes at both time points, and those related to immune function changed in people in their early 60s.
Abrupt changes in number
Snyder, the Stanford W. Ascherman, MD, FACS Professor in Genetics, and his colleagues were inspired to look at the rate of molecular and microbial shifts by the observation that the risk of developing many age-linked diseases does not rise incrementally along with years. For example, risks for Alzheimer’s disease and cardiovascular disease rise sharply in older age, compared with a gradual increase in risk for those under 60.
The researchers used data from 108 people they’ve been following to better understand the biology of aging. Past insights from this same group of study volunteers include the discovery of four distinct “ ageotypes ,” showing that people’s kidneys, livers, metabolism and immune system age at different rates in different people.

Michael Snyder
The new study analyzed participants who donated blood and other biological samples every few months over the span of several years; the scientists tracked many different kinds of molecules in these samples, including RNA, proteins and metabolites, as well as shifts in the participants’ microbiomes. The researchers tracked age-related changes in more than 135,000 different molecules and microbes, for a total of nearly 250 billion distinct data points.
They found that thousands of molecules and microbes undergo shifts in their abundance, either increasing or decreasing — around 81% of all the molecules they studied showed non-linear fluctuations in number, meaning that they changed more at certain ages than other times. When they looked for clusters of molecules with the largest changes in amount, they found these transformations occurred the most in two time periods: when people were in their mid-40s, and when they were in their early 60s.
Although much research has focused on how different molecules increase or decrease as we age and how biological age may differ from chronological age, very few have looked at the rate of biological aging. That so many dramatic changes happen in the early 60s is perhaps not surprising, Snyder said, as many age-related disease risks and other age-related phenomena are known to increase at that point in life.
The large cluster of changes in the mid-40s was somewhat surprising to the scientists. At first, they assumed that menopause or perimenopause was driving large changes in the women in their study, skewing the whole group. But when they broke out the study group by sex, they found the shift was happening in men in their mid-40s, too.
“This suggests that while menopause or perimenopause may contribute to the changes observed in women in their mid-40s, there are likely other, more significant factors influencing these changes in both men and women. Identifying and studying these factors should be a priority for future research,” Shen said.
Changes may influence health and disease risk
In people in their 40s, significant changes were seen in the number of molecules related to alcohol, caffeine and lipid metabolism; cardiovascular disease; and skin and muscle. In those in their 60s, changes were related to carbohydrate and caffeine metabolism, immune regulation, kidney function, cardiovascular disease, and skin and muscle.
It’s possible some of these changes could be tied to lifestyle or behavioral factors that cluster at these age groups, rather than being driven by biological factors, Snyder said. For example, dysfunction in alcohol metabolism could result from an uptick in alcohol consumption in people’s mid-40s, often a stressful period of life.
The team plans to explore the drivers of these clusters of change. But whatever their causes, the existence of these clusters points to the need for people to pay attention to their health, especially in their 40s and 60s, the researchers said. That could look like increasing exercise to protect your heart and maintain muscle mass at both ages or decreasing alcohol consumption in your 40s as your ability to metabolize alcohol slows.
“I’m a big believer that we should try to adjust our lifestyles while we’re still healthy,” Snyder said.
The study was funded by the National Institutes of Health (grants U54DK102556, R01 DK110186-03, R01HG008164, NIH S10OD020141, UL1 TR001085 and P30DK116074) and the Stanford Data Science Initiative.
- Rachel Tompa Rachel Tompa is a freelance science writer.
About Stanford Medicine
Stanford Medicine is an integrated academic health system comprising the Stanford School of Medicine and adult and pediatric health care delivery systems. Together, they harness the full potential of biomedicine through collaborative research, education and clinical care for patients. For more information, please visit med.stanford.edu .
Hope amid crisis
Psychiatry’s new frontiers


IMAGES
COMMENTS
2. Prepare your paper for submission. Download our get published quick guide opens in new tab/window, which outlines the essential steps in preparing a paper.(This is also available in Chinese opens in new tab/window).It is very important that you stick to the specific "guide for authors" of the journal to which you are submitting.
Step 1: Choosing a journal. Choosing which journal to publish your research paper in is one of the most significant decisions you have to make as a researcher. Where you decide to submit your work can make a big difference to the reach and impact your research has. It's important to take your time to consider your options carefully and ...
Free 1 hour monthly How to Get Published webinars cover topics including writing an article, navigating the peer review process, and what exactly it means when you hear "open access.". Join fellow researchers and expert speakers live, or watch our library of recordings on a variety of topics. Browse our webinars.
Sun and Linton (2014), Hierons (2016) and Craig (2010) offer useful discussions on the subject of "desk rejections.". 4. Make a good first impression with your title and abstract. The title and abstract are incredibly important components of a manuscript as they are the first elements a journal editor sees.
3. Submit your article according to the journal's submission guidelines. Go to the "author's guide" (or similar) on the journal's website to review its submission requirements. Once you are satisfied that your paper meets all of the guidelines, submit the paper through the appropriate channels.
To Publish a Research Paper follow the guide below: Conduct original research: Conduct thorough research on a specific topic or problem. Collect data, analyze it, and draw conclusions based on your findings. Write the paper: Write a detailed paper describing your research.
Open access is a publishing model in which the author pays a fee to publish; the reader is able to access the article for free. Some journals are entirely open access, while others are "hybrid"—providing both a subscription as well as an open access publishing option. Open science, on the other hand, is a movement towards increased ...
Choosing the journal before you start writing also means you can tailor your work to build on research that's already been published there. This can help editors to see how a paper adds to the 'conversation' in their journal. To help you with this crucial step, look at our guide on selecting the right journal for your research.
A. Yes, instead of giving the volume and page number, you can give the paper's DOI at the end of the citation. For example, Nature papers should be cited in the form; Author (s) Nature advance ...
Call for papers in your subject area. Journals regularly 'call for papers', asking for submissions within a particular field or topic. Answering these is a great way to get published, making sure your research fits the journal's aims and scope. Simply select your subject area with our handy tool to get started. Search current calls for ...
The introduction section should be approximately three to five paragraphs in length. Look at examples from your target journal to decide the appropriate length. This section should include the elements shown in Fig. 1. Begin with a general context, narrowing to the specific focus of the paper.
Communicating research findings is an essential step in the research process. Often, peer-reviewed journals are the forum for such communication, yet many researchers are never taught how to write a publishable scientific paper. In this article, we explain the basic structure of a scientific paper and describe the information that should be included in each section. We also identify common ...
An academic who is trying to get a journal article published is a lot like a salmon swimming upstream, says Dana S. Dunn, PhD, a member of APA's Board of Educational Affairs. ... a measure of how often papers in the journal are cited compared to how much is published in the journal. For more narrowly focused research, there are journals ...
Step 2: Finding the Right Journal. Understanding how to publish a research paper involves selecting the appropriate journal for your work. This step is critical for successful publication, and you should take several factors into account when deciding which journal to apply for: Conduct thorough research to identify journals that specialise in ...
In Tips for Publishing in Scientific Journals , Science Deputy Editor Katrina Kelner takes a peek into the publishing process and offers nuts-and-bolts advice on how to get your research into print. Roberta Ness, a widely published epidemiologist and a less widely published author of children's books, lets us in on a secret common to both types ...
How to Get Published in a Journal: 12 Proven Strategies for Academic Success. Increase your chances of academic publishing success! Learn insider tips for crafting strong research, choosing the right journal, and navigating the peer-review process. ... Starting a PhD with a goal to publish a research paper could be both exciting and daunting ...
Free 1 hour monthly How to Get Published webinars cover topics including writing an article, navigating the peer review process, and what exactly it means when you hear "open access.". Join fellow researchers and expert speakers live, or watch our library of recordings on a variety of topics. Browse our webinars.
The publisher sends proofs, and usually an order blank for reprints, to the author and asks that the materials be returned in 24 to 48 hours. Some journals send manuscripts to the publisher as soon as the article is accepted. In this case, the manuscript is typeset immediately and placed in a queue for publication.
Here are a few steps that you can take to significantly improve your chances of getting published: 1. Browse legit journals. As of 2015, the academic publishing market had an annual revenue of $20.5 Billion. This revenue has grown tremendously over the last two years.
Make an impact and build your research profile in the open with ScienceOpen. Search and discover relevant research in over 95 million Open Access articles and article records; Share your expertise and get credit by publicly reviewing any article; Publish your poster or preprint and track usage and impact with article- and author-level metrics; Create a topical Collection to advance your ...
STEM Fellowship Journal (SFJ) SFJ is a peer-reviewed journal published by Canadian Science Publishing that serves as a platform for scholarly research conducted by high school and university students in the STEM fields. Peer review is conducted by undergraduate, graduate student, and professional reviewers.
Elsevier Journal Finder helps you find journals that could be best suited for publishing your scientific article. Journal Finder uses smart search technology and field-of-research specific vocabularies to match your paper's abstract to scientific journals.
Find the research you need | With 160+ million publications, 1+ million questions, and 25+ million researchers, this is where everyone can access science
Scientific and technical journal publications per million residents of the world as of 2013. Academic publishing is the subfield of publishing which distributes academic research and scholarship. Most academic work is published in academic journal articles, books or theses.The part of academic written output that is not formally published but merely printed up or posted on the Internet is ...
Overall, this research demonstrates that functions and risks of aging-related diseases change nonlinearly across the human lifespan and provides insights into the molecular and biological pathways ...
Key Points. Question Are glucagon-like peptide-1 receptor agonists semaglutide and liraglutide, which were originally introduced for the treatment of type 2 diabetes and are frequently prescribed due to their weight loss properties, associated with disproportionately increased reporting of suicidality?. Findings This disproportionality analysis through the case-control design based on the ...
Strong et al. demonstrate the application of ionic electro-active polymer hydrogels to computational tasks through use of a multi-electrode array integrated with a simulated Pong environment, taking advantage of the unique memory mechanics present in many active mediums. Improved performance is shown within the task as environmental memory is acquired.
This Special Issue seeks for the latest research, novelties, and applications of FAS and other NGRA-enabled wireless communication technologies in 6G networks. We solicit original and high-quality papers that cover several topics of interest, including but not limited to:
A paper describing these findings was published in the journal Nature Aging Aug. 14. ... Identifying and studying these factors should be a priority for future research," Shen said. Changes may influence health and disease risk. In people in their 40s, significant changes were seen in the number of molecules related to alcohol, caffeine and ...
It discusses the potential of the UK's Modern Slavery Act 2015, despite its criticisms, and explores if such a reform could benefit Hong Kong. This study aims to stimulate dialogue and research to enhance legal frameworks combating human trafficking.