An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- BioTechnologia (Pozn)
- v.103(1); 2022


CRISPR/Cas9 in plant biotechnology: applications and challenges
CRISPR – clustered regularly interspaced short palindromic repeats
Cas9 – CRISPR-associated protein 9
GMO – genetically modified organism
PBT – plant biotechnology
SSN – sequence-specific nucleases
TALENs – transcription activator-like effector nucleases
ZFNs – Zinc finger nucleases
The application of plant biotechnology to enhance beneficial traits in crops is now indispensable because of food insecurity due to increasing global population and climate change. The recent biotechnological development of the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated system 9 (Cas9) allows for a more simple and precise method of gene editing, which is now preferred compared to Zinc Finger Nucleases (ZFNs) and Transcription Activator-like Effector Nucleases (TALENs). In this review, recent progress in utilizing CRISPR/Cas9-mediated gene editing in plants to enhance certain traits in beneficial crops, including rice, soybean, and oilseed rape, is discussed. In addition, novel methods of applying the CRISPR/Cas9 system in live cell imaging are also extensively reviewed. Despite all the applications, the existing delivery methods of CRISPR/Cas9 fail to provide consistent results and are inefficient for in planta transformation. Hence, research should be focused on improving current delivery methods or developing novel ones to facilitate CRISPR/Cas9-based gene editing studies. Strict regulations on the sale and commercial growth of gene-edited crops have restricted more efforts in applying CRISPR/Cas9 technology in plant species. Therefore, a shift in public viewpoint toward gene editing would help to propel scientific progress rapidly.
Introduction
With the rising demand of food security due to the ever-increasing population growth coupled with a looming threat of climate change (Haque et al. 2018 ; United Nations 2019 ), the urgency to develop reliable and efficient methods to secure a steady and sufficient nutrition to the global population is higher than ever. Hence, the role and application of plant biotechnology to engineer plants to suit global agricultural demands are now indispensable. Plant biotechnology (PBT), in essence, comprises the set of scientific methods and techniques used to identify and manipulate plant genes in order to develop desired traits or specific products in plants (Kalia, 2018 ). By using the methods available in PBT, beneficial traits of crops can be expressed and amplified, while undesirable traits and components such as allergens in rice, peanuts, or soybeans can be eliminated (Fuchs and Mackey, 2003 ; Barh and Azevedo, 2018 ).
The emergence of Sequence-Specific Nucleases (SSNs) such as Zinc Finger Nucleases (ZFNs), Transcriptional Activator-like Effector Nucleases (TALENs), and the more recently developed Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) are among the advanced methods that allowed a less sporadic and more precise means of genetic modifications in plants (Baltes et al., 2014 ; Fauser et al., 2014 ; Endo et al., 2016 ; Li et al., 2016 ; Sun et al., 2016a ; Sun et al., 2016b ). A detailed review on the comparison between the abovementioned three methods was done by Sun et al. ( 2016a ). The CRISPR/Cas9 system is generally preferred over the other methods because of its precision, efficiency, simplicity, and cost-effectiveness. Hence, it has gained attention in the genome editing community (Haque et al., 2018 ; Wang et al., 2018 ). Since the discovery of the first CRISPR locus by Ishino et al. ( 1987 ) and the pioneering extensive study on the CRISPR/Cas system by Jansen et al. ( 2002 ), the technology has been further studied for genome editing of various organisms.
In essence, CRISPR/Cas9 technology exploits the adaptive immunity system of the bacteria Streptococcus pyogenes in DNA repair to modify the genetic sequences or even edit the genome of the targeted organism. This is achieved by constructing a single guide RNA (sgRNA) specific to the target DNA sequence, which forms a complex with the Cas9 protein, thereby initiating specific doublestranded breaks in the target DNA, as shown in Fig. 1 (Costa et al., 2017 ). The double-stranded breaks enable further gene editing as shown in Figure 1D and Figure 1E . Many studies have been conducted to describe the mechanism of the CRISPR/Cas9 system, as were extensively reviewed by researchers (Sander and Joung, 2014 ; Westra et al., 2014 ; Bortesi and Fischer, 2015 ; Ma et al., 2015 ; Ma et al., 2016 ; Musunuru, 2017 ; Adhikari and Poudel, 2020 ). Hence, to follow suit, this review discusses 1) the recent advances in utilizing the CRISPR/Cas9 system in a diverse range of plant species for crop enhancement and facilitate plant cell imaging, 2) the current challenges faced regarding the delivery methods of CRISPR/Cas9 reagents into plant cells, and 3) the regulatory systems of gene edited crops compared to those for genetically modified organisms (GMOs).

Mechanism of CRISPR/Cas9 gene editing. A) The constructed target-specific single guide RNA (sgRNA) forms a complex with the Cas9 protein; B) The CRISPR/Cas9 complex binds to the target DNA; C) The CRISPR/Cas9 cleaves the target DNA at specific sequences, leading to further gene editing; D) Gene knock-in through homology-directed repair (HDR); E) Gene knock-out through non-homologous end joining (NHEJ)
Applications of CRISPR/Cas9 genome editing in plant species
Certain phenotypes or traits that are expressed by plants, or in this case crops, can be tweaked and adjusted through the manipulation of their genes. In doing so, the expected outcome would be to produce an enhanced version of the crop, which can be beneficial to the general population from certain aspects. The precision of CRISPR/Cas9 technology ensures a highly reliable method in genome editing that does not randomly produce unforeseen alterations elsewhere in the genome (Schiml et al., 2016 ). The efforts in trying to apply CRISPR/Cas9 genome editing in plants have been widespread since the discovery of the technology. Prior to applying the genome editing technology to crops, much of the research was conducted on Arabidopsis thaliana as a model plant organism because of its convenience and usefulness in genetic experiments (Koornneef and Meinke, 2010 ; Lee et al., 2018 ).
For example, A. thaliana was used as a model plant in implementing a sequential transformation method, which improved CRISPR gene targeting (Miki et al., 2018 ). The efficiency of pKAMA-ITACHI Red vector in CRISPR/Cas9 was also first investigated in A. thaliana when a study involving genes such as PDS3, AG , and DUO1 , was conducted by Tsutsui and Higashiyama ( 2017 ). After the initial validation on A. thaliana , the potentials of the technology are being further explored in other plant species. Some examples of plants and crops that have been successfully manipulated using CRISPR/Cas9 technology during the recent years are outlined in Table 1 .
Examples of successful genome editing of plant species
| Plant | Gene(s) targeted | Traits | Method | References |
|---|---|---|---|---|
| Apple | MdDIPM4 | disease resistance | gene inactivation | Pompili et al. |
| Maize | | flowering time/plant height | gene knockout & overexpression | Li et al. |
| Muskmelon | albinism (CRISPR trial) | gene knockout | Hooghvorst et al. | |
| Oil palm | | disease resistance | base editing | Budiani et al. |
| Oilseed rape | herbicide resistance | base editing | Wu et al. | |
| | plant flowering | gene knockout/down | Jiang et al. 2018 | |
| Rice | | disease resistance thermotolerance grain length salt tolerance | gene knockou gene knockout & overexpression site directed mutagenesis gene knockout & overexpression | Kim et al. Guo et al. Usman et al. X. Zhang et al. 2020 |
| Soybean | | flowering time & regional adaptability | site directed mutagenesis | Cai et al., 2018; Cai et al., ; Wang et al., |
| | disease resistance | multiplex gene knockout | P. Zhang et al., 2020 | |
| Tobacco | hybrid lethality | frameshift mutation | Ma et al., | |
| Watermelon | albinism (CRISPR trial) | gene knockout | Tian et al., |
Improvement on quality of crops
One of the more impactful applications of CRISPR/Cas9 technology from the sustainability aspect is the ability of the genome editing tool to enhance the quality of agricultural products. Rice, a major food source for the global population (Fukagawa and Ziska, 2019 ), was first successfully manipulated using the CRISPR/Cas9 technology by Miao et al. ( 2013 ), who demonstrated the possibility of applying the system for targeted mutations in rice. Since this finding, many efforts have been channeled to elucidate the functions of individual genes and observe the effect of gene alterations in rice in the hope to apply the findings practically. An example would be a study by Guo et al. ( 2020 ), who used CRISPR/Cas9 to both induce overexpression and knockout the OsProDH gene in rice. The OsProDH gene encodes for a mitochondrial enzyme, proline dehydrogenase, which is responsible for the degradation of proline in rice. Proline plays a significant role in protecting plants from various biotic and abiotic stresses by inducing diverse physiological responses of the plants and by scavenging reactive oxygen species (ROS) (Hayat et al., 2012 ). It was found that mutation in OsProDH in rice resulted in the accumulation of proline, which in turn led to lower levels of ROS (Guo et al., 2020 ). Hence, by manipulating the OsProDH gene and subsequently the metabolism of proline, higher thermotolerance could be conferred onto rice (Guo et al., 2020 ).
On the other hand, salt-tolerant rice can be potentially achieved through the manipulation of the OsNAC45 gene (Yu et al., 2018 ; Zhang et al., 2020b ). Through the regulation of several other plant stress response genes ( OsCYP89G1 , OsDREB1F , OsEREBP2 , OsERF104 , OsPM1 , OsSAMDC2 , OsSIK1 ), OsNAC45 may be significant in regulating abscisic acid signal responses in rice, which could be the key to produce rice with increased salt tolerance (Zhang et al., 2020b ). In another study, CRISPR/Cas9 was used to elucidate the role of polygalacturonase in regulating the cell wall immune response through the gene OsPG1 (Cao et al., 2021 ). This not only deepens understanding of the role of cell wall integrity in plant immune response but also highlights the potential to exploit the cell wall physiology in conferring bacterial resistance. Other research studies used CRISPR/Cas9 in a similar manner with the aim of producing an observable effect either through gene overexpression or gene knockout in rice. Through these methods, various genes have been identified and successfully manipulated, for example, genes responsible for traits such as pigment (anthocyanin) content (Zheng et al., 2019 ; Hu et al., 2020 ), resistance to disease (bacterial blight and blast disease) (Zhou et al., 2015 ; Wang et al., 2016 ; Kim et al., 2019 ), and grain length (Li et al., 2020a; Usman et al., 2021 ). These findings prove that CRISPR/Cas9 technology is undoubtedly effective in manipulating traits in rice, and it is expected that these methods can be applied to generate more resilient, robust, and nutritious rice, which can drive global sustainability.
CRISPR/Cas9-based mutagenesis has also been successfully performed on other impactful plant species with significant mutation efficiency. The first application of CRISPR/Cas9 gene editing in soybean was conducted by Jacobs et al. (2015) where gene knockout was performed on the green fluorescent protein (GFP) gene. This pioneer work kickstarted numerous efforts in applying CRISPR/Cas9 gene editing in soybean. Han et al. ( 2019 ) utilized CRISPR/Cas9 to induce a targeted mutation in the E1 gene in controlling soybean flowering and found that the truncation of the E1 protein prevented the inhibition of the GmFT2a/5a gene, increased its expression, and led to an earlier flowering time under long-day (LD) conditions. This transformation led to the development of a photo-insensitive soybean variant, which is potentially suitable for the introduction of soybean in higher latitudes (Han et al., 2019 ).
Similarly, a study conducted by Cai et al. ( 2020 ) demonstrated the role of the GmFT2a/5a gene in soybean in regulating flowering times and yield under different photoperiods by comparing double-knockouts and overexpression of the gene using CRISPR/Cas9 technology. These findings collectively established the involvement of certain genes in soybean that may contribute to its adaptability in different environments and conditions. Furthermore, the GmFT2a/5a double-knockout mutants were found to produce a significantly higher amount of pods and seeds per plants as compared to the wild-type plant, despite having a longer flowering time (Cai et al., 2020 ). In addition, the overexpression of GmPRR37 was found to lengthen flowering time under LD conditions and was involved in downregulating the aforementioned GmFT2a/5a , which promotes flowering, and in upregulating GmFT1a that inhibits flowering, thereby contributing to the regional adaptability of soybean (Wang et al., 2020 ). From these results, soybean variants with a higher productivity can be bred and adapted to a more diverse environment. Triple knockouts of GmF3H1 , GmF3H2 , and GmFNSII-1 were effectively performed using a multiplex CRISPR/Cas9 system in soybean and resulted in an increase in isoflavone content within the plants that at the same time conferred enhanced resistance to the soybean mosaic virus (SMV) (Zhang et al., 2020a ). Several genome edits in soybean were successfully inherited to subsequent generations (Han et al., 2019 ; Zhang et al., 2020a ), indicating that selective breeding of CRISPR/Cas9-edited soybean could potentially generate beneficial novel crop variants. However, the inheritance of CRISPR/Cas9 mutations requires further studies as the efficiency of its occurrence is still rather sporadic.
Oilseed rape
Oilseed rape ( Brassica napus ), also known as rapeseed, is another impactful crop that is notable for the production of edible oils (Cartea et al., 2019 ). The success in CRISPR/Cas9-mediated mutagenesis of rapeseed was first reported by Yang et al. ( 2017 ) where 12 genes from four gene families ( BnaA9.RGA , BnaC9.RGA , BnaA6.RGA , and BnaC7.RGA from the BnaRGA family; BnaA9.FUL , BnaC2.FUL , and BnaC7FUL from the BnaFUL family; and BnaA2.DA2.1 , BnaA2.DA2.2 , BnaC6.DA2 , BnaC5.DA1 , and BnaA6.DA1 from the BnaDA2 and BnaDA1 families) were tested in the study. Subsequently, stable inheritance of the induced mutations by the following progeny was observed in the study, indicating the effectiveness of CRISPR/Cas9 in producing an enhanced variant of oilseed rape (Yang et al., 2017 ). Following this study, Jiang et al. (2018) successfully identified the role of the BnaSDG8.A and BnaSGD8.C genes in promoting the expression of histone 3 lysine 36 (H3K36) methyltransferase, consequently influencing floral transition in oilseed rape as well as mutating the aforementioned genes to produce an early flowering phenotype. In addition, silencing the BnSFAR4 and BnSFAR5 genes in CRISPR/Cas9-mediated double gene knockout could increase the seed oil content (SOC) in oilseed rape without affecting seed germination, vigor, and oil mobilization, as demonstrated by Karunarathna et al. ( 2020 ). In another study, CRISPR/Cas9-mediated cytosine base-editing (CBE) was used in mutating the BnALS1 gene by introducing a C to T conversion at the specific region (Wu et al., 2020 ). This mutation produced a mutant oilseed rape that could resist tribenuron-methyl, a herbicide commonly used against weeds (Wu et al., 2020 ). Hence, the development of herbicide resistance in oilseed rape will help farmers in weed management. Taken together, these findings help to drive the productivity and to simplify the management of oilseed rape crop.
Other crop species
Currently, CRISPR/Cas9 genome editing has been demonstrated to be successful on a number of influential crops such as maize (Liu et al., 2020 ; Li et al., 2020b), wheat (Hayta et al., 2019 ; Liu et al., 2020 ), and apples (Pompili et al., 2020 ), with a relatively high transformation efficiency (Haque et al., 2018 ; Adhikari and Poudel, 2020 ). The sequencing of novel plant genomes had widened the applications of CRISPR/Cas9 genome editing in testing higher number of genes in various plant species. CRISPR/Cas9 was recently reported to be effective in knocking out the phytoene desaturase gene in muskmelon ( CmPDS ), which is the first reported study to apply CRISPR/Cas9 genome editing on the species (Hooghvorst et al., 2019 ). The same PDS gene was also successfully knocked out to produce an albino phenotype in CRISPR/Cas9 genome editing pioneering studies on watermelon and apples (Nishitani et al., 2016 ; Tian et al., 2017 ). However, the rate of inheritance by the subsequent generations of transgenic plants could not be investigated through PDS gene knockout as the albino variants had low in vitro survival rates (Hooghvorst et al., 2019 ); hence, other genes should be targeted to determine the rate of inheritance of mutations in these plant species.
Targeted mutagenesis in sweet orange was achieved by Jia and Nian ( 2014 ), where a novel tool for delivering the CRISPR/Cas9 reagents was developed for citrus plants through the Xcc-facilitated agroinfiltration, and involved the use of Xanthomonas citri subsp. citri (Xcc) to infect the citrus plant. Knockout of the CsWRKY22 gene in Wanjincheng orange using CRISPR/Cas9 genome editing exhibited enhanced resistance toward citrus canker, a destructive disease in citrus plants caused by Xcc, thereby further establishing the efficacy of CRISPR/Cas9 technology in citrus (Wang et al., 2019 ). Similar enhancement of disease resistance was observed in apples where the successful CRISPR/Cas9-mediated gene knockout of MdDIPM4 conferred increased resistance to Erwinia amylovora , a bacterium that causes fire blight disease in apples (Pompili et al., 2020 ). Pompili et al. ( 2020 ) could successfully clear CRISPR/Cas9 reagents from the genome by using T-DNA removal, which reduced the chances of occurrence of unnecessary or off-target mutations. As a conclusion, CRISPR/Cas9 technology can be applied to a diverse range of plant species and can produce a multitude of effects expressed by the plants. It is expected that the benefits of CRISPR/Cas9-edited crops and products would be able to reach the consumers. This, however, comes with its own set of challenges, one of which will be discussed in the later sections.
Live cell CRISPR imaging
Conventional cellular imaging methods applied in subnuclear dynamics studies such as fluorescence in situ hybridization (FISH) (Langer-Safer et al., 1982 ; Schwarzacher and Heslop-Harrison, 1994 ; Wu et al., 2019 ) are limited by the need of cellular fixation and the heat denaturation step that influence chromatin structure and organization, consequently impeding temporal studies in plant cells (Kozubek et al., 2000 ; Boettiger et al., 2016 ; Dreissig et al., 2017 ). Live cell imaging in plants allows spatiotemporal organization of chromatin to be studied in greater detail, which may potentially deepen the understanding of various gene expression patterns. Novel approaches in live cellular imaging tend to use Zinc Fingers (ZFs) or Transcription Activator-like Effectors (TALEs), which are proteins that can be programmed to bind to specific DNA sequences (Qin et al., 2017 ; Wu et al., 2019 ). Even though ZFs and TALEs are more flexible than FISH, there are technical challenges that one has to face as complicated processes are involved in constructing a large array of ZFs and TALEs proteins (Qin et al., 2017 ) and in constructing their expression vectors capable of targeting multiple DNA sequences (Wu et al., 2019 ). The necessity of re-engineering TALEs in targeting to a new gene sequence is also time-consuming and labor-intensive (Khosravi et al., 2020 ).
In view of the limitations of ZFs, TALEs, and FISH, researchers are utilizing the CRISPR/Cas system to achieve a live cell imaging method with greater flexibility and to overcome the limitations of visualizing non-repetitive regions (Dreissig et al., 2017 ). In this most recent approach, the nuclease activity-deficient dead Cas9 (dCas9), which was shown to possess specific DNA binding ability without DNA alterations (Qi et al., 2013 ; Dominguez et al., 2016 ), is combined with a fluorescence protein (FP) to visualize telomeric repeats in live leaf cells of Nicotiana benthamiana . The study proved the usefulness of this method to observe DNA-protein interactions in live plant cells (Dreissig et al., 2017 ). Telomere repeats in Nicotiana tabacum were also successfully labeled by transiently expressing dCas9-FP, mediated by an Agrobacterium vector (Fujimoto and Matsunaga, 2017 ). A protocol on conducting live plant cell imaging using CRISPR/Cas9 from S. pyogenes and Staphylococcus aureus was developed by Khosravi et al. ( 2020 ), where a telomere-specific guide RNA was used to target the telomeric sequences in N. benthamiana . Through these initial findings, the CRISPR/Cas9 imaging system shows potential for further development in visualizing gene sequences with low repetition or low abundance. Simple and reliable imaging of chromatin spatiotemporal organization would also ease further research on gene expression at various stages of the plant cell cycle. dCas9 can also be applied in gene expression inhibition, transcriptional regulation, gene promoter activation and for monitoring spatiotemporal patterns of gene expression in plants (Bikard and Marraffini, 2013 ; Yang, 2015 ; Arora and Narula, 2017 ). This shows that studies on a single system may potentially yield outcomes that can be beneficial and applied to multiple areas of interest. The potential of the CRISPR/Cas9 system has barely been explored, and more is yet to come.
Challenges in applying CRISPR/Cas9 technology in plants
As a relatively novel toolbox for genome editing, there are certainly some obstacles to be resolved when trying to apply CRISPR/Cas9 technology in plants. First, before any manipulation can be performed on the genome, the specific gene responsible for the intended function must be identified to enable precise editing. Despite the efforts conducted to sequence the genomes of many relevant plant species, there is still insufficient knowledge on the function of sequenced genes within the plants’ genome, which impedes efforts in precision editing to produce intended effects (Haque et al., 2018 ; Adhikari and Poudel, 2020 ). Fortunately, by conducting Genome-Wide Association Studies (GWAS), gene functions can be effectively predicted with accuracy, which can drive further research on necessary manipulations in plants. For instance, Zheng et al. ( 2019 ) discovered the genes OsC1 and OsRb that are involved in regulating anthocyanins in rice leaf. This enabled Hu et al. ( 2020 ) to further use the CRISPR/Cas9 technology in manipulating anthocyanin levels in rice. A similar approach was also undertaken to study the RDP1 gene of A. thaliana (Tsuchimatsu et al., 2020 ). Just as how GWAS can propel CRISPR/Cas9 plant editing, CRISPR/Cas9 technology is also used as an alternative method for cross population validation (Alseekh et al., 2021 ), such as to validate GWAS findings in rice ( Oryza sativa ) (Lu et al., 2017 ; Meng et al., 2017 ) and maize ( Zea mays ) (Liu et al., 2020 ). This provides an insight into the importance of establishing the causal relationships and interactions between genes that can further drive the development of CRISPR/Cas9 technology in plants (Yin et al., 2017 ).
Delivery and disposal of CRISPR/Cas9 reagents in plants
The delivery process of the necessary CRISPR/Cas9 components into intended cells remains a challenge to its application in plant and animal cells alike, especially in an in vivo setting (Li et al., 2015 ). Agrobacterium -mediated delivery using A. tumefaciens or A. rhizogenes is a commonly used method for plant transformation in various species (Ron et al., 2014 ; Mikami et al., 2015 ; Budiani et al., 2018 ; Hooghvorst et al., 2019 ; Mao et al., 2019 ; Pompili et al., 2020 ; Li et al., 2020b). Despite its popularity, there is still a degree of uncertainty when utilizing this method as its success depends on the choice of the plasmid and the cultivar used (Mangena et al., 2017 ). Various studies have reported that the A. rhizogenes -mediated transformation system could have been the cause of low transformation efficiency observed in soybean (Li et al., 2019 ; Bai et al., 2020 ; Zhang et al., 2020a ), rice (Butt et al., 2017 ; Usman et al., 2021 ), and tomato (Ron et al., 2014 ) genome editing. Varying culture conditions can also influence the infection and regeneration rates of the Agrobacterium- infected explants, which affects the reproducibility of the results obtained (Hamada et al., 2018 ) as observed in soybean (Li et al., 2017; Hada, 2018 ; Mangena, 2018 ), clover ( Trifolium subterraneum L.) (Rojo, 2021 ), and cassava (Nyaboga et al., 2015 ). This further showed inconsistencies observed in transformation efficiency through Agrobacterium mediated delivery. In addition, while a high degree of success was observed in A. thaliana , the feasibility of Agrobacterium -mediated transformation in other plant species such as soybean (Mangena et al., 2017 ), melon (Hooghvorst et al., 2019 ), and wheat (Zhang et al., 2018 ) is still questionable, where the regeneration of transgenic plants would require the use of explant-derived calluses (Mao et al., 2019 ). Hence, further studies are required to enhance the Agrobacterium -mediated delivery method to increase its transformation efficiency, effectiveness in diverse plant species, and its success for in planta transformation.
An alternative to the Agrobacterium -mediated delivery system is biolistic delivery (Carter and Shieh, 2015 ). Biolistic delivery is the direct delivery of DNA material into plant cells, where DNA is coated onto heavy metal particles such as gold or tungsten (Baltes et al., 2017 ). As the DNA-coated metal particles penetrate and get trapped inside plant cells, DNA can dissociate from the particles and become integrated into the host genome (Baltes et al., 2017 ). Although recent success in inducing in planta genome manipulation was observed in wheat ( Triticum aestivum L.), the mutation efficiency that was reported using the biolistic method remains very low, less than 6% of samples being mutated and less than 2% of samples with the mutations inherited (Hamada et al., 2018 ).
Another alternative involves the use of viral vectors as a delivery system for the CRISPR/Cas9 components. A study by Ma et al. ( 2020 ) utilizing the sonchus yellow net rhabdovirus (SYNV) to infect tobacco plants reported relatively high mutation efficiency with minimal costs, but the disadvantage of using viral vectors lies in the range of infectivity of the proposed virus. Nonetheless, reverse genetic tools can aid in expanding the range of infectivity for other rhabdoviruses (Ma et al., 2020 ). Hence, in planta genome editing using CRISPR/Cas9 is currently limited by the availability of effective delivery systems, and further studies and development of conventional and novel delivery methods would contribute to efficient research of CRISPR/Cas9 in plants.
CRISPR/Cas9-edited crop regulation
The ultimate goal of developing novel methods and innovations in applying the CRISPR/Cas9 technology in PBT is to enhance the quality of life of consumers through the production of transgenic plants or crops. Gene-edited organisms such as the ones edited using CRISPR/Cas9 technology involve mutagenesis of their genomes through either deletions, substitutions, or insertions of base pairs, while GMOs involve the introduction of a foreign genetic material or transgene into the organism that may or may not be integrated into the genome (Callaway, 2018 ). Despite this fundamental difference, gene-edited organisms are often governed by the same set of rules and regulations as those for GMOs in many countries (El-Mounadi et al., 2020 ). For instance, the Court of Justice of the European Union (CJEU) had recently ruled that gene-edited crops are not exempted by laws and regulations governing GM crops (Callaway, 2018 ; Confédération paysanne and others v. Premier ministre and Ministre de l’Agriculture de l’Agroalimentaire et de la Forêt, 2018 ). This implies that the high hurdles that were put in place in developing GM crops also apply to CRISPR/Cas9-edited crops, which may drive funding and investment away from future research on CRISPR/Cas9 as a viable plant breeding technology. The EU’s unchanging definition of GMOs as “not naturally altered” further impacted the public perception toward CRISPR technology and genetic modification as a whole (Plan and Eede, 2010 ). The road to gain public confidence toward GMOs on their safety, efficacy, and benefits is already riddled with various aspects of social, economic, and legal challenges (Zimny et al., 2019 ). However, shifting the public perspective toward gene technology is the key to trigger much needed changes across the board.
In contrast to the EU, the US Department of Agriculture (USDA) ruled out regulation of genome-edited plants, provided its production does not involve plant pests (USDA, 2018 ). In addition to highlighting the safety and the lack of risks involved with genome-edited plants, this new ruling would promote further progress in the development of the technology (Hoffman, 2021 ). The first of the genome-edited crops allowed to bypass USDA regulations is a CRISPR/Cas9-edited white button mushroom resistant to browning (Waltz, 2016 ). The USDA has also been continuously funding research involving CRISPR-edited plants such as rice ( O. sativa ) (Lee et al., 2019 ), pennycress ( Thlaspi arvense L.) (Jarvis et al., 2021 ), and cocoa ( Theobroma cacao ) (Fister et al., 2018 ). Integrating modern technological approaches into regulations that were designed for older technology cannot possibly be the way forward. In contrast, law and regulations require modernization to keep up with the transformative power of innovation. Hence, rather than treating old GMO regulations as an umbrella that cannot continuously cover new and upcoming technologies such as CRISPR, regulations need to be amended as necessary.
However, despite periodic updates in GMO regulations and the development of novel guidelines, Malaysia is yet to approve the commercial growth of genome-edited crops (Singh et al., 2019 ). Similar to EU, Malaysia’s regulatory system classified genome-edited crops under GMOs; hence, any plant or crops would be difficult to gain approval by the system (El-Mounadi et al., 2020 ). Although Malaysia is relatively reserved in approving gene-edited crop propagation in the open field, it allowed more than 30 cases of import of transgenic products, albeit solely for the purpose of consumption or processing, in addition to approving confined field tests of transgenic plants such as rubber and papaya (Singh et al., 2019 ).
To be fair, crops produced by CRISPR/Cas9 gene editing and other gene editing methods utilized globally challenge the conventional perspectives and definition of gene modification and GMOs. Hence, there is no doubt that the regulatory bodies worldwide are still adapting to the rapid development of this technology. Therefore, despite legal hurdles, researchers, investors, and consumers alike should retain their interests in the development and research of more beneficial crops so that the supply would be able to cope with the rise in food demand.
Conclusions
CRISPR/Cas9 has received much attention in recent years as a revolutionary technology to genetically manipulate organisms to suit our demands. While initial research and development studied were focused on animal cell lines, the utilization of CRISPR/Cas9 gene editing has now been expanded to be inclusive of a diverse range of plant species, specifically beneficial and important crops. Through the enhancement of agricultural crops, agricultural and nutrition demands are expected to be met in an effort to improve the global quality of life. It has also been shown that the potential of CRISPR/Cas9 is not limited to the improvement of phenotypical traits, as this technology can also be used in live plant cell imaging to facilitate scientific research. There could be additional new methods to exploit CRISPR/Cas9 in the coming years, and this development should be anticipated in the fast-paced modernized era of scientific innovation. Therefore, scientific progress should not be discouraged or even impeded by issues concerning outdated regulation systems. This, coupled with low public acceptance and valuation of GMOs and CRISPR in general (Shew et al., 2018 ), indirectly influence the availability of funding toward further research. However, with patience and collaborative efforts from scientific community in sharing the knowledge and presenting advances in practical aspects of science, a shift in public perspective toward not just CRISPR/Cas9 but gene editing as a whole, would help to propel rapidly scientific progress in genome editing.
This review did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interest
The authors declare that there is no conflict of interest.
Acknowledgements
The authors wish to thank Prof. Hoe I. Ling of Columbia University (New York, USA) for his editorial input.
- Adhikari P., Poudel M. (2020) CRISPR-Cas9 in agriculture: Approaches, applications, future perspectives, and associated challenges . Malays. J. Halal Res . 3 ( 1 ): 6–16. 10.2478/mjhr-2020-0002 [ CrossRef ] [ Google Scholar ]
- Alseekh S., Kostova D., Bulut M., Fernie A.R. (2021) Genome-wide association studies: assessing trait characteristics in model and crop plants . Cell. Mol. Life Sci . 78 ( 15 ): 5743–5754. 10.1007/S00018-021-03868-W [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Arora L., Narula A. (2017) Gene editing and crop improvement using CRISPR-cas9 system . Front. Plant Sci . 8 :1932. 10.3389/fpls.2017.01932 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Bai M., Yuan J., Kuang H., Gong P., Li S., Zhang Z., Liu B., Sun J., Yang M., Yang L., et al.. (2020) Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean . Plant Biotechnol J . 18 ( 3 ): 721–731. 10.1111/PBI.13239 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Baltes N.J., Gil-Humanes J., Cermak T., Atkins P.A., Voytas D.F. (2014) DNA replicons for plant genome engineering . Plant Cell . 26 ( 1 ): 151–163. 10.1105/TPC.113.119792 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Baltes N.J., Gil-Humanes J., Voytas D.F. (2017) Genome engineering and agriculture: opportunities and challenges. Chapter 1 . [In:] Progress in molecular biology and translational science. Vol. 149 . Ed. Weeks D.P., Yang B., San Diego: Academic Press: 1–26. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Barh D., Azevedo V. (2018) Omics technologies and bio-engineering. Volume 2: towards improving quality of life . London: Elsevier. [ Google Scholar ]
- Bikard D., Marraffini L.A. (2013) Control of gene expression by CRISPR-Cas systems . F1000Prime Rep . 5 : 47. 10.12703/P5-47 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Boettiger A.N., Bintu B., Moffitt J.R., Wang S., Beliveau B.J., Fudenberg G., Imakaev M., Mirny L.A., Wu C.T., Zhuang X. (2016) Super-resolution imaging reveals distinct chromatin folding for different epigenetic states . Nature . 529 ( 7586 ): 418–422. 10.1038/nature16496 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Bortesi L., Fischer R. (2015) The CRISPR/Cas9 system for plant genome editing and beyond . Biotechnol. Adv . 33 ( 1 ): 41–52. 10.1016/j.biotechadv.2014.12.006 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Budiani A., Putranto R.A., Riyadi I., Sumaryono, Minarsih H.,Faizah R. (2018) Transformation of oil palm calli using CRISPR/Cas9 System: Toward genome editing of oil palm . IOP Conf. Ser. Earth Environ. Sci . 183 : 12003. [ Google Scholar ]
- Butt H., Eid A., Ali Z., Atia M.A.M., Mokhtar M.M., Hassan N., Lee C.M., Bao G., Mahfouz M.M. (2017) Efficient CRISPR/Cas9-mediated genome editing using a chimeric single-guide RNA molecule . Front Plant Sci . 0 : 1441. 10.3389/FPLS.2017.01441 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Cai Y., Wang L., Chen L., Wu T., Liu L., Sun S., Wu C., Yao W., Jiang B., Yuan S., et al.. (2020) Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean . Plant Biotechnol. J . 18 ( 1 ): 298–309. 10.1111/pbi.13199 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Callaway E. (2018) CRISPR plants now subject to tough GM laws in European Union . Nature . 560 ( 7716 ): 16. 10.1038/d41586-018-05814-6 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Cao Y., Zhang Y., Chen Y., Yu N., Liaqat S., Wu W., Chen D., Cheng S., Wei X., Cao L., et al.. (2021) OsPG1 encodes a olygalacturonase that determines cell wall architecture and affects resistance to bacterial blight pathogen in rice . Rice . 14 ( 1 ): 1–15. 10.1186/S12284-021-00478-9 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Cartea E., Haro-Bailón A. de, Padilla G., Obregón-Cano S., Rio-Celestino M.D., Ordás A. (2019) Seed oil quality of Brassica napus and Brassica rapa germplasm from Northwestern Spain . Foods . 8 ( 8 ): 292. 10.3390/FOODS8080292 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Carter M., Shieh J. (2015) Gene delivery strategies. Chapter 11 . [in:] Guide to Research Techniques in Neuroscience (Second Edition). London: Academic Press: 239–252. [ Google Scholar ]
- Confédération paysanne and others v. Premier ministre and Ministre de l’Agriculture de l’Agroalimentaire et de la Forêt . (2018) Judgment of the Court of Justice of the European Union in the case C-528/16 . Luxembourg. [accessed 2021 Sep 14]. https://curia.europa.eu/jcms/upload/docs/application/pdf/2018-07/cp180111en.pdf [ Google Scholar ]
- Costa J.R., Bejcek B.E., McGee J.E., Fogel A.I., Brimacombe K.R., Ketteler R. (2017) Genome editing using engineered nucleases and their use in genomic screening . [In:] Assay guidance manual . Ed. Markossian S., Grossman A., Brimacombe K., Bethesda: Eli Lilly & Company and the National Center for Advancing Translational Sciences. [ PubMed ] [ Google Scholar ]
- Dominguez A.A., Lim W.A., Qi L.S. (2016) Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation . Nat. Rev. Mol. Cell Biol . 17 ( 1 ): 5–15. 10.1038/nrm.2015.2 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Dreissig S., Schiml S., Schindele P., Weiss O., Rutten T., Schubert V., Gladilin E., Mette M.F., Puchta H., Houben A. (2017) Live-cell CRISPR imaging in plants reveals dynamic telomere movements . Plant J . 91 ( 4 ): 565–573. 10.1111/tpj.13601 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- El-Mounadi K., Morales-Floriano M.L., Garcia-Ruiz H. (2020) Principles, applications, and biosafety of plant genome editing using CRISPR-Cas9 . Front. Plant Sci . 11 : 56. 10.3389/FPLS.2020.00056 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Endo M., Mikami M., Toki S. (2016) Biallelic gene targeting in rice . Plant Physiol . 170 ( 2 ): 667–677. 10.1104/PP.15.01663 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Fauser F., Schiml S., Puchta H. (2014) Both CRISPR/Casbased nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana . Plant J . 79 ( 2 ): 348–359. 10.1111/TPJ.12554 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Fister A.S., Landherr L., Maximova S.N., Guiltinan M.J. (2018) Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao . Front. Plant Sci . 268. 10.3389/FPLS.2018.00268 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Fuchs R.L., Mackey M.A. (2003) Genetically modified foods . [In:] Encyclopedia of food sciences and nutrition , Second Edition Ed. Caballero B. Elsevier Science: 2876–2882. [ Google Scholar ]
- Fujimoto S., Matsunaga S. (2017) Visualization of chromatin loci with transiently expressed CRISPR/Cas9 in plants . Cytologia 82 ( 5 ): 559–562. 10.1508/cytologia.82.559 [ CrossRef ] [ Google Scholar ]
- Fukagawa N.K., Ziska L.H. (2019) Rice: importance for global nutrition . J. Nutr. Sci. Vitaminol . 65 ( Supplement ): S2–S3. 10.3177/JNSV.65.S2 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Guo M., Zhang X., Liu J., Hou L., Liu H., Zhao X. (2020) OsProDH negatively regulates thermotolerance in rice by modulating proline metabolism and reactive oxygen species scavenging . Rice . 13 ( 1 ): 1–5. 10.1186/s12284-020-00422-3 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Hada A., Krishnan V., Mohamed Jaabir M.S., Kumari A., Jolly M., Praveen S., Sachdev A. (2018) Improved Agrobacterium tumefaciens-mediated transformation of soybean [Glycine max (L.) Merr.] following optimization of culture conditions and mechanical techniques . Vitr Cell Dev Biol-Plant . 54 ( 6 ): 672–688. 10.1007/S11627-018-9944-8 [ CrossRef ] [ Google Scholar ]
- Hamada H., Liu Y., Nagira Y., Miki R., Taoka N., Imai R. (2018) Biolistic-delivery-based transient CRISPR/Cas9 expression enables in planta genome editing in wheat . Sci. Rep . 8 ( 1 ): 14422. 10.1038/s41598-018-32714-6 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Han J., Guo B., Guo Y., Zhang B., Wang X., Qiu L.J. (2019) Creation of early flowering germplasm of soybean by CRISPR/Cas9 technology . Front. Plant Sci . 10 : 1446. 10.3389/fpls.2019.01446 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Haque E., Taniguchi H., Hassan M.M., Bhowmik P., Karim M.R., Śmiech M., Zhao K., Rahman M., Islam T. (2018) Application of CRISPR/Cas9 genome editing technology for the improvement of crops cultivated in tropical climates: Recent progress, prospects, and challenges . Front. Plant Sci . 9 : 617. 10.3389/fpls.2018.00617 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Hayat S., Hayat Q., Alyemeni M.N., Wani A.S., Pichtel J., Ahmad A. (2012) Role of proline under changing environments: A review . Plant Signal. Behav . 7 ( 11 ): 1456–1466. 10.4161/PSB.21949 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Hayta S., Smedley M.A., Demir S.U., Blundell R., Hinchliffe A., Atkinson N., Harwood W.A. (2019) An efficient and reproducible Agrobacterium-mediated transformation method for hexaploid wheat (Triticum aestivum L.) . Plant Meth . 15 ( 1 ): 1–15. 10.1186/S13007-019-0503-Z [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Hoffman N.E. (2021) Revisions to USDA biotechnology regulations: the SECURE rule . Proc. Natl. Acad. Sci. USA 118 ( 22 ): e2004841118. 10.1073/PNAS.2004841118 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Hooghvorst I., López-Cristoffanini C., Nogués S. (2019) Efficient knockout of phytoene desaturase gene using CRISPR/Cas9 in melon . Sci. Rep . 9 ( 1 ): 1–7. 10.1038/s41598-019-53710-4 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Hu W., Zhou T., Han Z., Tan C., Xing Y. (2020) Dominant complementary interaction between OsC1 and two tightly linked genes, Rb1 and Rb2, controls the purple leaf sheath in rice . Theor. Appl. Genet . 133 ( 9 ): 2555–2566. 10.1007/s00122-020-03617-w [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ishino Y., Shinagawa H., Makino K., Amemura M., Nakatura A. (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isoenzyme conversion in Escherichia coli, and identification of the gene product . J. Bacteriol . 169 ( 12 ): 5429–5433. 10.1128/jb.169.12.5429-5433.1987 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Jansen R., van Embden J.D.A., Gaastra W., Schouls L.M. (2002) Identification of genes that are associated with DNA repeats in prokaryotes . Mol. Microbiol . 43 ( 6 ): 1565–1575. 10.1046/j.1365-2958.2002.02839.x [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Jarvis B.A., Romsdahl T.B., McGinn M.G., Nazarenus T.J., Cahoon E.B., Chapman K.D., Sedbrook J.C. (2021) CRISPR/Cas9-induced fad2 and rod1 mutations stacked with fae1 confer high oleic acid seed oil in Pennycress (Thlaspi arvense L.) . Front. Plant Sci . 652. 10.3389/FPLS.2021.652319 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Jia H., Nian W. (2014) Targeted genome editing of sweet orange using Cas9/sgRNA . PLoS One . 9 ( 4 ): e93806. 10.1371/journal.pone.0093806 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Kalia A. (2018). Nanotechnology in bioengineering: transmogrifying plant biotechnology . [In:] Omics Technologies and Bio-engineering. Vol. 2: Towards Improving Quality of Life . Academic Press: 211–229. [ Google Scholar ]
- Karunarathna N.L., Wang H., Harloff H.J., Jiang L., Jung C. (2020) Elevating seed oil content in a polyploid crop by induced mutations in seed fatty acid reducer genes . Plant Biotechnol. J . 18 : 2251–2266. 10.1111/pbi.13381 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Khosravi S., Dreissig S., Schindele P., Wolter F., Rutten T., Puchta H., Houben A. (2020) Live-cell CRISPR imaging in plant cells with a telomere-specific guide RNA . Methods Mol. Biol . 2166 :343–356. 10.1007/978-10716-0712-1_20 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Kim Y.A., Moon H., Park C.J. (2019) CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae . Rice . 12 ( 1 ): 67. 10.1186/s12284-019-0325-7 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Koornneef M., Meinke D. (2010) The development of Arabidopsis as a model plant . Plant J . 61 ( 6 ): 909–921. 10.1111/j.1365-313X.2009.04086.x [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Kozubek S., Lukášová E., Amrichová J., Kozubek M., Lišková A., Šlotová J. (2000) Influence of cell fixation on chromatin topography . Anal. Biochem . 282 ( 1 ): 29–38. 10.1006/abio.2000.4538 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Langer-Safer P.R., Levine M., Ward D.C. (1982) Immunological methods for mapping genes on Drosophila polytene chromosomes . Proc. Natl. Acad. Sci. USA . 79 ( 14 ): 4381–4385. 10.1073/pnas.79.14.4381 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Lee K., Eggenberger A.L., Banakar R., McCaw M.E., Zhu H., Main M., Kang M., Gelvin S.B., Wang K. (2019) CRISPR/Cas9-mediated targeted T-DNA integration in rice . Plant Mol. Biol . 99 ( 4 ): 317–328. 10.1007/S11103-018-00819-1 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Lee Z.H., Yamaguchi N., Ito T. (2018) Using CRISPR/Cas9 system to introduce targeted mutation in Arabidopsis . Meth. Mol. Biol . 1830 : 93–108. 10.1007/978-1-4939-8657-6_6 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Li C., Nguyen V., Liu J., Fu W., Chen C., Yu K., Cui Y. (2019) Mutagenesis of seed storage protein genes in soybean using CRISPR/Cas9 . BMC Res. Notes . 12 ( 1 ): 176. 10.1186/s13104-019-4207-2 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Li L., He Z.Y., Wei X.W., Gao G.P., Wei Y.Q. (2015) Challenges in CRISPR/CAS9 delivery: potential roles of nonviral vectors . Hum. Gene Ther . 26 ( 7 ): 452–462. 10.1089/hum.2015.069 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Li Q., Lu L., Liu H., Bai X., Zhou X., Wu B., Yuan M., Yang L., Xing Y. (2020) A minor QTL, SG3, encoding an R2R3-MYB protein, negatively controls grain length in rice . Theor. Appl. Genet . 133 ( 8 ): 2387–2399. 10.1007/s00122-020-03606-z [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Li Q., Wu G., Zhao Y., Wang B., Zhao B., Kong D., Wei H., Chen C., Wang H. (2020) CRISPR/Cas9-mediated knockout and overexpression studies reveal a role of maize phytochrome C in regulating flowering time and plant height . Plant Biotechnol. J . 18 ( 12 ): 2520–2532. 10.1111/pbi.13429 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Li T., Liu B., Chen C.Y., Yang B. (2016) TALEN-mediated homologous recombination produces site-directed DNA base change and herbicide-resistant rice . J. Genet. Genomics . 43 ( 5 ): 297–305. 10.1016/j.jgg.2016.03.005 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Liu H., Wang K., Jia Z., Gong Q., Lin Z., Du L., Pei X., Ye X. (2020) Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system . J. Exp. Bot . 71 ( 4 ): 1337–1349. 10.1093/JXB/ERZ529 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Liu H.J., Jian L., Xu J., Zhang Q., Zhang M., Jin M., Peng Y., Yan J., Han B., Liu J. et al.. (2020) High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize . Plant Cell . 32 ( 5 ): 1397–1413. 10.1105/TPC.19.00934 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Lu Y., Ye X., Guo R., Huang J., Wang W., Tang J., Tan L., Zhu J., Chu C., Qian Y. (2017) Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system . Mol. Plant . 10 ( 9 ): 1242–1245. 10.1016/J.MOLP.2017.06.007 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., Wang B., Yang Z., Li H., Lin Y., et al.. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants . Mol. Plant . 8 ( 8 ): 1274–1284. 10.1016/j.molp.2015.04.007 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ma X., Zhang X., Liu H., Li Z. (2020) Highly efficient DNA-free plant genome editing using virally delivered CRISPR –Cas9 . Nat. Plants . 6 ( 7 ): 773–779. 10.1038/s41477-020-0704-5 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ma X., Zhu Q., Chen Y., Liu Y.G. (2016) CRISPR/Cas9 platforms for genome editing in plants: developments and applications . Mol. Plant . 9 ( 7 ): 961–974. 10.1016/j.molp.2016.04.009. [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Mangena P., Mokwala P.W., Nikolova R.V. (2017) Challenges of in vitro and in vivo Agrobacterium-mediated genetic transformation in soybean . [In:] Soybean – the basis of yield, biomass and productivity . Ed. Kasai M., InTech Open. [ Google Scholar ]
- Mangena P. (2018) The role of plant genotype, culture medium and Agrobacterium on soybean plantlets regeneration during genetic transformation . [In:] Transgenic Crops – Emerging Trends and Future Perspectives . Ed. Khan M.S., Malik K.A., InTechOpen. [ Google Scholar ]
- Mao Y., Botella J.R., Liu Y., Zhu J.K. (2019) Gene editing in plants: Progress and challenges . Natl. Sci. Rev . 6 ( 3 ): 421–437. 10.1093/nsr/nwz005 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Meng X., Yu H., Zhang Y., Zhuang F., Song X., Gao S., Gao C., Li J. (2017) Construction of a genome-wide mutant library in rice using CRISPR/Cas9 . Mol. Plant . 10 ( 9 ): 1238–1241. 10.1016/J.MOLP.2017.06.006 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Miao J., Guo D., Zhang J., Huang Q., Qin G., Zhang X., Wan J., Gu H., Qu L.J. (2013) Targeted mutagenesis in rice using CRISPR-Cas system . Cell Res . 23 ( 10 ): 1233–1236. 10.1038/cr.2013.123 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Mikami M., Toki S., Endo M. (2015) Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice . Plant Mol. Biol . 88 ( 6 ): 561–572. 10.1007/s11103-015-0342-x [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Miki D., Zhang W., Zeng W., Feng Z., Zhu J.K. (2018) CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation . Nat. Commun . 9 ( 1 ): 1967. 10.1038/S41467-018-04416-0. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Musunuru K. (2017) The hope and hype of CRISPR-Cas9 genome editing: A review . JAMA Cardiol . 2 ( 8 ): 914–919. 10.1001/jamacardio.2017.1713 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Nishitani C., Hirai N., Komori S., Wada M., Okada K., Osakabe K., Yamamoto T., Osakabe Y. (2016) Efficient genome editing in apple using a CRISPR/Cas9 system . Sci. Rep . 6 : 31481. 10.1038/srep31481 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Nyaboga E.N., Njiru J.M., Tripathi L. (2015) Factors influencing somatic embryogenesis, regeneration, and Agrobacterium-mediated transformation of cassava (Manihot esculenta Crantz) cultivar TME14 . Front Plant Sci . 6 :411. 10.3389/FPLS.2015.00411 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Plan D., Van Den Eede G. (2010) The EU Legislation on GMOs – an overview . EUR 24279 EN . Luxembourg (Luxembourg): Publications Office of the European Union; JRC57223. [ Google Scholar ]
- Pompili V., Dalla Costa L., Piazza S., Pindo M., Malnoy M. (2020) Reduced fire blight susceptibility in apple cultivars using a high-efficiency CRISPR/Cas9-FLP/FRT-based gene editing system . Plant Biotechnol. J . 18 ( 3 ): 845–858. 10.1111/pbi.13253 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression . Cell . 152 ( 5 ): 1173–1183. 10.1016/j.cell.2013.02.022 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Qin P., Parlak M., Kuscu C., Bandaria J., Mir M., Szlachta K., Singh R., Darzacq X., Yildiz A., Adli M. (2017) Live cell imaging of low- and non-repetitive chromosome loci using CRISPR-Cas9 . Nat. Commun . 8 ( 1 ): 1–10. 10.1038/ncomms14725 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Rojo F.P., Seth S., Erskine W., Kaur P. (2021) An improved protocol for Agrobacterium-mediated transformation in subterranean clover (Trifolium subterraneum l.) . Int. J. Mol. Sci . 22 ( 8 ): 4181. 10.3390/ijms22084181 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ron M., Kajala K., Pauluzzi G., Wang D., Reynoso M.A., Zumstein K., Garcha J., Winte S., Masson H., Inagaki S., et al.. (2014) Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model . Plant Physiol . 166 ( 2 ): 455–469. 10.1104/pp.114.239392 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Sander J.D., Joung J.K. (2014) CRISPR-Cas systems for editing, regulating and targeting genomes . Nature Biotechnol . 32 ( 4 ): 347–350. 10.1038/nbt.2842 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Schiml S., Fauser F., Puchta H. (2016) CRISPR/Cas-mediated site-specific mutagenesis in Arabidopsis thaliana using Cas9 nucleases and paired nickases . Methods Mol. Biol . 1469 : 111–122. 10.1007/978-1-4939-4931-1_8 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Schwarzacher T., Heslop-Harrison J.S. (1994) Direct fluorochrome-labeled DNA probes for direct fluorescent in situ hybridization to chromosomes . Methods Mol. Biol . 28 : 167–176. 10.1385/0-89603-254-x:167 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Singh D.J.K., Mat Jalaluddin N.S., Sanan-Mishra N., Harikrishna J.A. (2019) Genetic modification in Malaysia and India: current regulatory framework and the special case of non-transformative RNAi in agriculture . Plant Cell Rep . 38 ( 12 ): 1449–1463. 10.1007/s00299-019-02446-6 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Shew A.M., Nalley L.L., Snell H.A., Nayga R.M., Dixon B.L. (2018) CRISPR versus GMOs: Public acceptance and valuation . Glob. Food Sec . 19 : 71–80. 10.1016/J.GFS.2018.10.005 [ CrossRef ] [ Google Scholar ]
- Sun Y., Li J., Xia L. (2016a) Precise genome modification via sequence-specific nucleases-mediated gene targeting for crop improvement . Front. Plant Sci . 7 : 1928. 10.3389/fpls.2016.01928 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Sun Y., Zhang X., Wu C., He Y., Ma Y., Hou H., Guo X., Du W., Zhao Y., Xia L. (2016b) Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase . Mol. Plant . 9 ( 4 ): 628–631. 10.1016/J.MOLP.2016.01.001 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Tian S., Jiang L., Gao Q., Zhang J., Zong M., Zhang H., Ren Y., Guo S., Gong G., Liu F., et al.. (2017) Efficient CRISPR/Cas9-based gene knockout in watermelon . Plant Cell Rep . 36 ( 3 ): 399–406. 10.1007/s00299-016-2089-5 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Tsuchimatsu T., Kakui H., Yamazaki M., Marona C., Tsutsui H., Hedhly A., Meng D., Sato Y., Städler T., Grossniklaus U. et al.. (2020) Adaptive reduction of male gamete number in the selfing plant Arabidopsis thaliana . Nat. Commun . 11 ( 1 ): 1–9. 10.1038/s41467-020-16679-7 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Tsutsui H., Higashiyama T. (2017) pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana . Plant Cell Physiol . 58 ( 1 ): 46–56. 10.1093/PCP/PCW191. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- United Nations . (2019) World Population Prospects 2019 . [accessed 2020 Sep 7]. https://population.un.org/wpp/Download/Standard/Population/ .
- USDA . (2018) Secretary Perdue Issues USDA Statement on Plant Breeding Innovation . [accessed 2021 Sep 15]. https://www.usda.gov/media/press-releases/2018/03/28/secretary-perdue-issues-usda-statement-plant-breeding-innovation .
- Usman B., Zhao N., Nawaz G., Qin B., Liu F., Liu Y., Li R. (2021) CRISPR/Cas9 guided mutagenesis of grain size 3 confers increased rice (Oryza sativa L.) grain length by regulating cysteine proteinase inhibitor and ubiquitin-related proteins . Int. J. Mol. Sci . 22 ( 6 ): 1–19. 10.3390/IJMS22063225 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Waltz E. (2016) Gene-edited CRISPR mushroom escapes US regulation . Nature . 532 ( 7599 ): 293. 10.1038/nature.2016.19754 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wang F., Wang C., Liu P., Lei C., Hao W., Gao Y., Liu Y.G., Zhao K. (2016) Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922 . PLoS One . 11 ( 4 ): e0154027. 10.1371/journal.pone.0154027 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wang L., Chen S., Peng A., Xie Z., He Y., Zou X. (2019) CRISPR/Cas9-mediated editing of CsWRKY22 reduces susceptibility to Xanthomonas citri subsp. citri in Wanjin-cheng orange (Citrus sinensis (L.) Osbeck) . Plant Biotechnol. Rep . 13 ( 5 ): 501–510. 10.1007/s11816-019-00556-x [ CrossRef ] [ Google Scholar ]
- Wang L., Sun S., Wu T., Liu L., Sun X., Cai Y., Li J., Jia H., Yuan S., Chen L. et al.. (2020) Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean . Plant Biotechnol. J . 18 ( 9 ): 1869–1881. 10.1111/pbi.13346 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wang M., Wang S., Liang Z., Shi W., Gao C., Xia G. (2018) From genetic stock to genome editing: gene exploitation in wheat . Trends Biotechnol . 36 ( 2 ): 160–172. 10.1016/j.tibtech.2017.10.002 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Westra E.R., Buckling A., Fineran P.C. (2014) CRISPR-Cas systems: Beyond adaptive immunity . Nature Rev. Microbiol . 12 ( 5 ): 317–326. 10.1038/nrmicro3241 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wu J., Chen C., Xian G., Liu D., Lin L., Yin S., Sun Q., Fang Y., Zhang H., Wang Y. (2020) Engineering herbicide-resistant oilseed rape by CRISPR/Cas9-mediated cytosine base-editing . Plant. Biotechnol. J . 18 ( 9 ): 1857–1859. 10.1111/pbi.13368 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wu X., Mao S., Ying Y., Krueger C.J., Chen A.K. (2019) Progress and challenges for live-cell imaging of genomic loci using CRISPR-based platforms . Genomics Proteomics Bioinformatics . 17 ( 2 ): 119–128. 10.1016/j.gpb.2018.10.001 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Yang H., Wu J.J., Tang T., Liu K.D., Dai C. (2017) CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus . Sci. Rep . 7 ( 1 ): 1–13. 10.1038/s41598-017-07871-9 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Yang X. (2015) Applications of CRISPR-Cas9 mediated genome engineering . Mil. Med. Res . 2 ( 1 ): 11. 10.1186/s40779-015-0038-1 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Yin K., Gao C., Qiu J.L. (2017) Progress and prospects in plant genome editing . Nat. Plants . 3 : 17107. 10.1038/nplants.2017.107 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Yu S., Huang A., Li J., Gao L., Feng Y., Pemberton E., Chen C. (2018) OsNAC45 plays complex roles by mediating POD activity and the expression of development-related genes under various abiotic stresses in rice root . Plant Growth Regul . 84 ( 3 ): 519–531. 10.1007/S10725-017-0358 [ CrossRef ] [ Google Scholar ]
- Zhang P., Du H., Wang .J, Pu Y., Yang C., Yan R., Yang H., Cheng H., Yu D. (2020a) Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus . Plant Biotechnol. J . 18 ( 6 ): 1384–1395. 10.1111/pbi.13302 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhang S., Zhang R., Song G., Gao J., Li W., Han X., Chen M., Li Y., Li G. (2018) Targeted mutagenesis using the Agrobacterium tumefaciens-mediated CRISPR-Cas9 system in common wheat . BMC Plant Biol . 18 ( 1 ): 1–12. 10.1186/S12870-018-1496-X [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhang X., Long Y., Huang J., Xia J. (2020b) OsNAC45 is involved in ABA response and salt tolerance in rice . Rice . 13 ( 1 ): 1–13. 10.1186/s12284-020-00440-1 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zheng J., Wu H., Zhu H., Huang C., Liu C., Chang Y., Kong Z., Zhou Z., Wang G., Lin Y., et al.. (2019) Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves . New Phytol . 223 ( 2 ): 705–721. 10.1111/nph.15807 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhou J., Peng Z., Long J., Sosso D., Liu B., Eom J.S., Huang S., Liu S., Vera Cruz C., Frommer W.B., et al.. (2015) Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice . Plant J . 82 ( 4 ): 632–643. 10.1111/tpj.12838 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zimny T., Sowa S., Tyczewska A., Twardowski T. (2019) Certain new plant breeding techniques and their marketability in the context of EU GMO legislation – recent developments . New Biotechnol . 51 : 49–56. 10.1016/J.NBT.2019.02.003 [ PubMed ] [ CrossRef ] [ Google Scholar ]

- General & Introductory Agriculture
Plant Biotechnology Journal
Print ISSN: 1467-7644
Online ISSN: 1467-7652
Impact Factor: 10.1
Association of Applied Biologists , Society for Experimental Biology
Edited By:Johnathan Napier

Recent Advances in Plant Biotechnology
- © 2009
- Ara Kirakosyan 0 ,
- Peter B. Kaufman 1
University of Michigan, Ann Arbor, U.S.A.
You can also search for this author in PubMed Google Scholar
- Presents a full overview of plant biotechnology from the history to applications
- Approach includes associated risks and the effects of plant biotechnology on global warming, alternative energy initiatives, food production, and medicine
- Includes supplementary material: sn.pub/extras
33k Accesses
81 Citations
4 Altmetric
This is a preview of subscription content, log in via an institution to check access.
Access this book
Subscribe and save.
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
- Available as EPUB and PDF
- Read on any device
- Instant download
- Own it forever
- Compact, lightweight edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
- Durable hardcover edition
Tax calculation will be finalised at checkout
Other ways to access
Licence this eBook for your library
Institutional subscriptions
About this book
Similar content being viewed by others.

Plant Genetics and Molecular Biology: An Introduction

History of Plant Biotechnology Development

- agriculture
- alternative energy
- bioremediation
- biotechnology
- genetically modified plants
- herbal medicine
- herbal products
- plant biotechnology
- transgenic plants
Table of contents (16 chapters)
Front matter, plant biotechnology from inception to the present, overview of plant biotechnology from its early roots to the present.
- Ara Kirakosyan, Peter B. Kaufman, Leland J. Cseke
The Use of Plant Cell Biotechnology for the Production of Phytochemicals
- Ara Kirakosyan, Leland J. Cseke, Peter B. Kaufman
Molecular Farming of Antibodies in Plants
- Rainer Fischer, Stefan Schillberg, Richard M. Twyman
Use of Cyanobacterial Proteins to Engineer New Crops
- Matias D. Zurbriggen, Néstor Carrillo, Mohammad-Reza Hajirezaei
Molecular Biology of Secondary Metabolism: Case Study for Glycyrrhiza Plants
- Hiroaki Hayashi
Applications of Plant Biotechnology in Agriculture and Industry
New developments in agricultural and industrial plant biotechnology, phytoremediation: the wave of the future.
- Jerry S. Succuro, Steven S. McDonald, Casey R. Lu
Biotechnology of the Rhizosphere
- Beatriz Ramos Solano, Jorge Barriuso Maicas, Javier Gutierrez Mañero
Plants as Sources of Energy
- Leland J. Cseke, Gopi K. Podila, Ara Kirakosyan, Peter B. Kaufman
Use of Plant Secondary Metabolites in Medicine and Nutrition
Interactions of bioactive plant metabolites: synergism, antagonism, and additivity.
- John Boik, Ara Kirakosyan, Peter B. Kaufman, E. Mitchell Seymour, Kevin Spelman
The Use of Selected Medicinal Herbs for Chemoprevention and Treatment of Cancer, Parkinson’s Disease, Heart Disease, and Depression
- Maureen McKenzie, Carl Li, Peter B. Kaufman, E. Mitchell Seymour, Ara Kirakosyan
Regulating Phytonutrient Levels in Plants – Toward Modification of Plant Metabolism for Human Health
Risks and benefits associated with plant biotechnology, risks and benefits associated with genetically modified (gm) plants.
- Peter B. Kaufman, Soo Chul Chang, Ara Kirakosyan
Risks Involved in the Use of Herbal Products
- Peter B. Kaufman, Maureen McKenzie, Ara Kirakosyan
Risks Associated with Overcollection of Medicinal Plants in Natural Habitats
- Maureen McKenzie, Ara Kirakosyan, Peter B. Kaufman
Authors and Affiliations
Ara Kirakosyan, Peter B. Kaufman
About the authors
Bibliographic information.
Book Title : Recent Advances in Plant Biotechnology
Authors : Ara Kirakosyan, Peter B. Kaufman
DOI : https://doi.org/10.1007/978-1-4419-0194-1
Publisher : Springer New York, NY
eBook Packages : Biomedical and Life Sciences , Biomedical and Life Sciences (R0)
Copyright Information : The Editor(s) (if applicable) and The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature 2009
Hardcover ISBN : 978-1-4419-0193-4 Published: 30 July 2009
Softcover ISBN : 978-1-4899-7916-2 Published: 23 August 2016
eBook ISBN : 978-1-4419-0194-1 Published: 15 August 2009
Edition Number : 1
Number of Pages : XIV, 405
Topics : Plant Genetics and Genomics , Plant Sciences
- Publish with us
Policies and ethics
- Find a journal
- Track your research

An official website of the United States government
Here's how you know
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
About Grants
The lifecycle of grants and cooperative agreements consists of four phases: Pre-Award, Award, Post-Award, and Close Out.
Access to Data
The National Institute of Food and Agriculture is committed to serving its stakeholders, Congress, and the public by using new technologies to advance greater openness.

Access Data Gateway
The Data Gateway enables users to find funding data, metrics, and information about research, education, and Extension projects that have received grant awards from NIFA.
View Resources Page
This website houses a large volume of supporting materials. In this section, you can search the wide range of documents, videos, and other resources.

Featured Webinar
Second annual virtual grants support technical assistance workshop.
Check out this five-day workshop in March 2024 workshop, designed to help you learn about NIFA grants and resources for grants development and management.
The National Institute of Food and Agriculture provides leadership and funding for programs that advance agriculture-related sciences.

Plant Biotechnology
Plant biotechnology is a set of techniques used to adapt plants for specific needs or opportunities. Situations that combine multiple needs and opportunities are common.
For example, a single crop may be required to provide sustainable food and healthful nutrition, protection of the environment, and opportunities for jobs and income. Finding or developing suitable plants is typically a highly complex challenge.
Plant biotechnologies that assist in developing new varieties and traits include genetics and genomics , marker-assisted selection (MAS) , and transgenic (genetic engineered) crops. These biotechnologies allow researchers to detect and map genes, discover their functions, select for specific genes in genetic resources and breeding, and transfer genes for specific traits into plants where they are needed. NIFA funds research, training, and extension for developing and using biotechnologies for food and agriculture. Areas of work include, but not limited to:
- Genetic structures and mechanisms
- Methods for transgenic biotechnology (also known as genetic engineering)
- Identification of traits and genes that can contribute to national and global goals for agriculture
- Plant genome sequences; molecular markers, and bioinformatics
- Gene Editing/Genome Editing
- Synthetic Biology
Most public research on transgenic crops focuses in some way on two general objectives:
- Better understanding of all aspects of the transgenic/genetic engineering process, for enhancing efficiency, precision, and proper expression of the added genes or nucleic acid molecules.
- A wider range of useful and valuable traits, including complex traits.
Competitive Funding Programs in Plant Biotechnology
Agriculture and Food Research Initiative (AFRI) program
Biotechnology Risk assessment Grants (BRAG) program
Small Business Innovation Research (SBIR) program
Specialty Crop Research Initiative (SCRI) program
Examples of Biotechnology Risk Assessment Grant (BRAG) projects
Latest Updates
- Latest Funding Opportunities
- Latest Blogs
- Latest Impacts
funding opportunity
Community food projects competitive grants program, specialty crop research initiative, air force 4-h military partnership outreach support grant program, hsis strengthening the nation’s workforce, usda nifa invests $300,000 to enhance forest and rangeland sustainability (rrea-nnf), usda nifa invests $2.5 million in regional rural development centers (rrdc), best radicchio varieties for new england farmers, beginning farmer and rancher development program participants report increased income, revenue and profits, multistate research projects address avian influenza, your feedback is important to us..
- Biomedical Science
- Biotechnological Engineering
- Plant Biotechnology
The future of plant biotechnology in a globalized and environmentally endangered world
- January 2020
- Genetics and Molecular Biology 43(1)

- Ghent University
Discover the world's research
- 25+ million members
- 160+ million publication pages
- 2.3+ billion citations

- Jovanka Miljuš-Đukić

- Edoardo Lenci

- Debra J Skinner
- Venkatesan Sundaresan

- NAT BIOTECHNOL
- Agustin Zsögön

- Andrew Balmford

- Mariana Mazzucato
- Calestous Juma

- FIELD CROP RES

- Juliette Legler
- Recruit researchers
- Join for free
- Login Email Tip: Most researchers use their institutional email address as their ResearchGate login Password Forgot password? Keep me logged in Log in or Continue with Google Welcome back! Please log in. Email · Hint Tip: Most researchers use their institutional email address as their ResearchGate login Password Forgot password? Keep me logged in Log in or Continue with Google No account? Sign up
Our websites may use cookies to personalize and enhance your experience. By continuing without changing your cookie settings, you agree to this collection. For more information, please see our University Websites Privacy Notice .
UConn Today
September 12, 2024 | Anna Zarra Aldrich, College of Agriculture, Health and Natural Resources
New Research on Plant Stem Cells Shines Light on How Plants Grow Stronger
We shouldn't ignore the potential of stem cells in plants, according to new UConn research

(Francesco Gallarotti for Unsplash)
Stem cell research is a hot topic. With applications for a host of human medical advancements, researchers have been working with animal and human stem cells for years.
But animals aren’t the only ones with stem cells.
Huanzhong Wang, professor of plant molecular biology in the College of Agriculture, Health and Natural Resources (CAHNR), wants people to know that plants have stem cells too. Just like in the medical world, plant stem cells could support human growth and development when used to improve the food supply.
“It’s not just humans and animals,” Wang says. “Plants have stem cells too, and we should be paying attention to them.”
In their roots, shoots, and vasculature, stem cells control cell division and differentiation for plants. Plant stem cells play a vital role in growth and development.
“Plants can grow for many, many years because different types of stem cells basically ensure they can grow up in the air and deep into the ground,” Wang says. “To grow a thicker stem or trunk, they need another type of stem cell.”
Plant stem cells have largely been overlooked because they don’t have applications for human biomedical research. But that doesn’t make them any less fascinating. And Wang has demonstrated that better understanding how these cells work can support a more resilient food supply.
Wang’s lab has been working with plant stem cells for years trying to understand how they control their stem cells, specifically the stem cells that give rise to vascular bundles – the structures that carry water and other nutrients throughout the plant.
Recently the group published a paper in New Phytologist that sheds light on this question.
Wang’s lab discovered a transcription factor gene called HVA that controls cell division in vascular stem cells.
When this gene is overexpressed, the researchers observed an increase in the number of vascular bundles and overall stem cell activity.
The researchers compared plants with no overexpression of HVA gene, those with one copy of overexpressed HVA gene and one regular gene, and finally plants with two copies of overexpressed HVA genes.
In the group with no overexpression, the plants had five to eight vascular bundles. In the plants with one copy of overexpressed HVA gene, they had more than 20 bundles, and with two copies of overexpressed HVA genes they had more than 50.
Aside from advancing science’s understanding of how plants work, Wang’s findings have important implications for agriculture.
Plants with more vascular bundles are stronger and more resistant to wind. This knowledge could be used to intentionally generate sturdier cultivars with the overexpression mutation.
This is especially relevant for tall, slender crops like corn, the biggest crop for the U.S.
“When plants grow taller, there is a risk that they could topple over,” Wang says. “Having more vascular bundles ensures the plant can stand still and resist those conditions.”
Even though Wang’s lab conducted the study using a model organism in the mustard family, the HVA gene is found in other plants as well, making this research broadly applicable.
HVA is one of hundreds of transcription factors in a large family in the plant’s genome. Wang is interested in discovering what some of the other genes in this family do.
“We are interested in studying other closely related genes to find out their function,” Wang says. “It will be interesting to study further how this gene family affects vascular development.
This work relates to CAHNR’s Strategic Vision area focused on Ensuring a Vibrant and Sustainable Agricultural Industry and Food Supply.
Follow UConn CAHNR on social media
Recent Articles

September 12, 2024
Connecting for Sickle Cell
Read the article

Science in Seconds – Eating Less, Living Longer

UConn to Host Workshop on Digital Twins for Manufacturing
share this!
September 12, 2024
This article has been reviewed according to Science X's editorial process and policies . Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
New research on plant stem cells shines light on how plants grow stronger
by Anna Zarra Aldrich, University of Connecticut

Stem cell research is a hot topic. With applications for a host of human medical advancements, researchers have been working with animal and human stem cells for years.
But animals aren't the only ones with stem cells.
Huanzhong Wang, professor of plant molecular biology in the College of Agriculture, Health and Natural Resources (CAHNR), wants people to know that plants have stem cells too. Just like in the medical world, plant stem cells could support human growth and development when used to improve the food supply.
"It's not just humans and animals," Wang says. "Plants have stem cells too, and we should be paying attention to them."
In their roots, shoots, and vasculature, stem cells control cell division and differentiation for plants. Plant stem cells play a vital role in growth and development.
"Plants can grow for many, many years because different types of stem cells basically ensure they can grow up in the air and deep into the ground," Wang says. "To grow a thicker stem or trunk, they need another type of stem cell."
Plant stem cells have largely been overlooked because they don't have applications for human biomedical research. But that doesn't make them any less fascinating. And Wang has demonstrated that better understanding how these cells work can support a more resilient food supply.
Wang's lab has been working with plant stem cells for years trying to understand how they control their stem cells, specifically the stem cells that give rise to vascular bundles—the structures that carry water and other nutrients throughout the plant.
Recently the group published a paper in New Phytologist that sheds light on this question. Wang's lab discovered a transcription factor gene called HVA that controls cell division in vascular stem cells.
When this gene is overexpressed, the researchers observed an increase in the number of vascular bundles and overall stem cell activity.
The researchers compared plants with no overexpression of HVA gene, those with one copy of overexpressed HVA gene and one regular gene, and finally plants with two copies of overexpressed HVA genes.
In the group with no overexpression, the plants had five to eight vascular bundles. In the plants with one copy of the overexpressed HVA gene, they had more than 20 bundles, and with two copies of overexpressed HVA genes they had more than 50.
Aside from advancing science's understanding of how plants work, Wang's findings have important implications for agriculture.
Plants with more vascular bundles are stronger and more resistant to wind. This knowledge could be used to intentionally generate sturdier cultivars with the overexpression mutation.
This is especially relevant for tall, slender crops like corn, the biggest crop in the U.S.
"When plants grow taller, there is a risk that they could topple over," Wang says. "Having more vascular bundles ensures the plant can stand still and resist those conditions."
Even though Wang's lab conducted the study using a model organism in the mustard family, the HVA gene is found in other plants as well, making this research broadly applicable.
HVA is one of hundreds of transcription factors in a large family in the plant's genome. Wang is interested in discovering what some of the other genes in this family do.
"We are interested in studying other closely related genes to find out their function," Wang says. "It will be interesting to study further how this gene family affects vascular development.
Journal information: New Phytologist
Provided by University of Connecticut
Explore further
Feedback to editors

Quantum researchers cause controlled 'wobble' in the nucleus of a single atom
11 minutes ago

Spectroscopic study unveils key steps for turning CO₂ into valuable chemicals
12 minutes ago

Respiratory biology study finds zebrafish use tastebuds to measure oxygen levels in water
37 minutes ago

Fundamental spintronics research reveals generic approach to magnetic second-order topological insulators

Laser and X-ray combo creates star-like conditions inside a hair-thin wire
38 minutes ago

New map shows where landslides are most likely to occur in US
58 minutes ago

Antarctica's receding sea ice could impact seabirds' food supply

Powered by renewable energy, microbes turn CO₂ into protein and vitamins

From Mount Etna to the UK: Genetics unveil the Oxford ragwort unique journey and resilience


An unprecedented feat: Printing 3D photonic crystals that completely block light
Relevant physicsforums posts, why does a series of pulses generate a pitch.
2 hours ago
Epothilone B study connected to 'Hard Problem of Consciousness' Model
Sep 9, 2024
Any stereo audio learning resources for other languages?
Sep 8, 2024
Too much fluoride might lower IQ in kids?
Sep 6, 2024
The predictive brain (Stimulus-Specific Error Prediction Neurons)
Sep 1, 2024
Any suggestions to dampen the sounds of a colostomy bag?
Aug 31, 2024
More from Biology and Medical
Related Stories

'Conductor' gene found in plant root stem cell 'orchestra'
Dec 6, 2019
Study sheds light on stem cell proliferation that may one day boost crop yields
Aug 2, 2018

A new mechanism behind continuous stem cell activity in plants
Jun 10, 2021
Recreating development in a petri dish to understand how plants live
Mar 21, 2024

A matter of concentration
Sep 17, 2019

How plant stem cells guard against genetic damage
Aug 27, 2021
Recommended for you

New in-vitro technique provides glimpse into the chloroplast workshop

Killer toxins produced by yeast may help remedy a craft beer brewing bother
3 hours ago

Parasitoid wasp that lays its eggs inside of adult fruit fly discovered
Let us know if there is a problem with our content.
Use this form if you have come across a typo, inaccuracy or would like to send an edit request for the content on this page. For general inquiries, please use our contact form . For general feedback, use the public comments section below (please adhere to guidelines ).
Please select the most appropriate category to facilitate processing of your request
Thank you for taking time to provide your feedback to the editors.
Your feedback is important to us. However, we do not guarantee individual replies due to the high volume of messages.
E-mail the story
Your email address is used only to let the recipient know who sent the email. Neither your address nor the recipient's address will be used for any other purpose. The information you enter will appear in your e-mail message and is not retained by Phys.org in any form.
Newsletter sign up
Get weekly and/or daily updates delivered to your inbox. You can unsubscribe at any time and we'll never share your details to third parties.
More information Privacy policy
Donate and enjoy an ad-free experience
We keep our content available to everyone. Consider supporting Science X's mission by getting a premium account.
E-mail newsletter
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
- My Bibliography
- Collections
- Citation manager
Save citation to file
Email citation, add to collections.
- Create a new collection
- Add to an existing collection
Add to My Bibliography
Your saved search, create a file for external citation management software, your rss feed.
- Search in PubMed
- Search in NLM Catalog
- Add to Search
Single-cell transcriptomics is revolutionizing the improvement of plant biotechnology research: recent advances and future opportunities
Affiliations.
- 1 Division of Research and Development, Plant Biotechnology Lab, Lovely Professional University, Phagwara, Punjab, India.
- 2 Department of Biotechnology, Lovely Faculty of Technology and Sciences, Lovely Professional University, Phagwara, Punjab, India.
- 3 Department of Research Facilitation, Division of Research and Development, Lovely Professional University, Phagwara, Punjab, India.
- 4 Agroécologie, InstitutAgro Dijon, INRAE, Univ. Bourgogne Franche-Comté, Dijon, France.
- PMID: 36775666
- DOI: 10.1080/07388551.2023.2165900
Single-cell approaches are a promising way to obtain high-resolution transcriptomics data and have the potential to revolutionize the study of plant growth and development. Recent years have seen the advent of unprecedented technological advances in the field of plant biology to study the transcriptional information of individual cells by single-cell RNA sequencing (scRNA-seq). This review focuses on the modern advancements of single-cell transcriptomics in plants over the past few years. In addition, it also offers a new insight of how these emerging methods will expedite advance research in plant biotechnology in the near future. Lastly, the various technological hurdles and inherent limitations of single-cell technology that need to be conquered to develop such outstanding possible knowledge gain is critically analyzed and discussed.
Keywords: Plants; plant transcriptomics; scRNA-seq; single-cell technology.
PubMed Disclaimer
Publication types
- Search in MeSH
LinkOut - more resources
Full text sources.
- Taylor & Francis
Miscellaneous
- NCI CPTAC Assay Portal
- Citation Manager
NCBI Literature Resources
MeSH PMC Bookshelf Disclaimer
The PubMed wordmark and PubMed logo are registered trademarks of the U.S. Department of Health and Human Services (HHS). Unauthorized use of these marks is strictly prohibited.
https://defraenvironment.blog.gov.uk/2024/09/12/a-call-for-plant-health-experts-to-support-defras-research/
A call for plant health experts to support Defra’s research

My name’s Alex and I am a scientist in the Plant Health Evidence and Analysis team. We use plant health research to fill any gaps in evidence and to inform Defra’s policies.
This month , the Defra Plant Health team launched a new ‘Register of Reviewers’ . This will provide the team with a list of external plant health experts, whom the team can approach when they need people with specific knowledge to review new research.
Independent reviews lead to better research – and ultimately better policy
An independent review by a relevant expert, also known as ‘peer review’, is an important part of research and evidence gathering.
Peer review is used to assess and assure the quality of research, whether it is value for money, and its reliability. To do this, they will carefully check that the project methods, results and outputs are scientifically sound and reliable.
This process holds researchers accountable for producing credible work and promotes research integrity. It ensures that research uses appropriate methodology, and that high-quality research is published.
At Defra, independent reviews help us to ensure that our research proposals are appropriately designed, and that our projects are good value for money.
And, ultimately, peer reviews mean that we can be confident that we’re using robust and reliable evidence to inform our decision-making and policies.
There are several opportunities for plant health peer reviewers
There is a vast range of plant health research happening across Defra, to support the department’s priorities. The main themes for our research are:
- risk assessment and horizon scanning, to help better prepare for the potential arrival of pests and diseases, and to inform our preventative regulation
- i nspections, diagnostics, and surveillance to help identify and prevent pests
- pest and disease m anagement to remove or reduce risk , and allow plants to recover
- r esilience and adaptation , which are key to the long-term health and sustainability of our ecosystems
- p lant health behaviours , to understand human behaviour around plant health and biosecurity, and to improve our knowledge of the economic, environmental, and cultural values of healthy plants
- developing evaluation frameworks and metrics to help us measure the effects of our plant health policies and work
For example, recent Defra research in plant health has included an Ash tree restoration project – the ‘Living Ash Project’. This project has helped to identify thousands of trees that are resistant to Ash dieback, a disease that has devastated Ash populations in the UK, to support a breeding programme of resistant Ash trees.

Being a peer reviewer has multiple benefits
Acting as a peer reviewer has benefits for the reviewer as well as for Defra. Taking part gives you the opportunity to:
- st ay up to date with current research , since reviewers will be assessing new and emerging research
- support impactful research by ensuring it is scientifically robust
- establish your presence and demonstrate your credibility in the field
- improve your skills by critically evaluating reports and proposals , then applying the same best - practice techniques to your own work
If you are interested in joining our register, you can find more information and apply here.
Sharing and comments
Share this page, leave a comment.
Cancel reply
By submitting a comment you understand it may be published on this public website. Please read our privacy notice to see how the GOV.UK blogging platform handles your information.
Related content and links
About this blog.
This blog is managed by Defra’s Nature Communications team.
On this blog, we share stories about our environmental work.
We cover topics including nature, water and biosecurity.
Find out more
Sign up and manage updates
- Defra on LinkedIn
- Defra Nature on Twitter
- Natural England on LinkedIn
- Natural England on Twitter
Using this blog
Read our guidelines

Research Specialist Seeds Team
- Madison, Wisconsin
- COLLEGE OF AGRICULTURAL & LIFE SCIENCES/WISCONSIN CROP INNOVATION CENTER
- Staff-Full Time
- Opening at: Sep 11 2024 at 11:05 CDT
- Closing at: Sep 26 2024 at 23:55 CDT
Job Summary:
The Wisconsin Crop Innovation Center (WCIC) at the University of Wisconsin-Madison is a ~100,000 square foot state-of-the-art agricultural biotechnology complex, housing laboratories and greenhouses dedicated to the production and analysis of transgenic and gene edited plants. This facility is the largest publicly held plant transformation facility in the United States and is aimed at improving crop plants for humans, livestock, and the environment. This position will focus on helping with the seeds team, one of eight teams at the WCIC. The Seeds team focuses on cataloging the material as it is harvested, and storing it for shipping, reporting what seeds are available and working with the clients to ensure they get their material out of storage when it is ready. Some clients will move material back to the greenhouse for growing to future generations. This position will also work to support the project coordinator for updating clients on their research progress. The College of Agricultural and Life Sciences (CALS) is committed to maintaining and growing a culture that embraces diversity, inclusion, and equity, believing that these values are foundational elements of our excellence and fundamental components of a positive and enriching learning and working environment for all students, faculty, and staff. At CALS, we acknowledge that bias, prejudice, racism, and hate have historically occurred in many forms that cause significant and lasting harm to members of our community. We commit to taking actions each day toward a college that is inclusive and welcoming to all.
Responsibilities:
- 30% Conducts research experiments according to established research protocols with moderate impact to the project(s). Collects data and monitors test results
- 10% Operates, cleans, and maintains organization of research equipment and research area. Tracks inventory levels and places replenishment orders
- 20% Reviews, analyzes, and interprets data and/or documents results for presentations and/or reporting to internal and external audiences
- 20% Participates in the development, interpretation, and implementation of research methodology and materials
- 10% Provides operational guidance on day-to-day activities of unit or program staff and/or student workers
- 10% Performs literature reviews and writes reports
Institutional Statement on Diversity:
Diversity is a source of strength, creativity, and innovation for UW-Madison. We value the contributions of each person and respect the profound ways their identity, culture, background, experience, status, abilities, and opinion enrich the university community. We commit ourselves to the pursuit of excellence in teaching, research, outreach, and diversity as inextricably linked goals. The University of Wisconsin-Madison fulfills its public mission by creating a welcoming and inclusive community for people from every background - people who as students, faculty, and staff serve Wisconsin and the world. For more information on diversity and inclusion on campus, please visit: Diversity and Inclusion
Preferred Bachelor's Degree
Qualifications:
Required: -Experience working in a lab environment handling and maintaining of samples for storage and later retrieval Preferred: -Genetics-based experience (coursework, lab work, field work) -Strong organizational skills -Experience with documentation and data entry
Full Time: 100% It is anticipated this position requires work be performed in-person, onsite, at a designated campus work location.
Appointment Type, Duration:
Ongoing/Renewable
Minimum $45,000 ANNUAL (12 months) Depending on Qualifications The minimum salary range for this position is $45,000. However, final salary will depend on experience and qualifications. Employees in this position can expect to receive benefits such as generous vacation, holidays, and paid time off; competitive insurances and savings accounts; retirement benefits. Additional benefits information can be found at: https://www.wisconsin.edu/ohrwd/benefits/download/fasl.pdf
Additional Information:
-Criminal Background Check may be required -A 12-month Evaluation Period may be required -The successful applicant will be responsible for ensuring eligibility for employment in the United States by the start of the appointment. University sponsorship is not available for this position.
How to Apply:
Click on the "Apply Online" button to start the application process. You will be prompted to upload the following documents/Application Materials: Resume (required) - Detail your educational and professional background Cover letter (required) - Refer to your related work experience References (required) - List contact information for three (3) references, including your current/most recent supervisor. References will not be contacted without prior notice. It's important that your cover letter and resume reflect your experience for this position related to the Qualifications section. Your application materials will be used during our evaluation to determine your qualifications as they relate to the job. The most qualified applicants will be invited to participate in the next step of the selection process.
Justin Cave [email protected] 608-263-3715 Relay Access (WTRS): 7-1-1. See RELAY_SERVICE for further information.
Official Title:
Research Specialist(RE047)
Department(s):
A07-COL OF AG & LIFE SCIENCES/WCIC/WCIC
Employment Class:
Academic Staff-Renewable
Job Number:
The university of wisconsin-madison is an equal opportunity and affirmative action employer..
You will be redirected to the application to launch your career momentarily. Thank you!
Frequently Asked Questions
Applicant Tutorial
Disability Accommodations
Pay Transparency Policy Statement
Refer a Friend
You've sent this job to a friend!
Website feedback, questions or accessibility issues: [email protected] .
Learn more about accessibility at UW–Madison .
© 2016–2024 Board of Regents of the University of Wisconsin System • Privacy Statement
Before You Go..
Would you like to sign-up for job alerts.
Thank you for subscribing to UW–Madison job alerts!
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
Biotechnology articles from across Nature Portfolio
Biotechnology is a broad discipline in which biological processes, organisms, cells or cellular components are exploited to develop new technologies. New tools and products developed by biotechnologists are useful in research, agriculture, industry and the clinic.

Recoded gene circuits for multiplexed genetic code expansion
Quadruplet codons allow multiplexing of non-canonical amino acids within single polypeptides in living cells. We show that including high-usage triplet codons after quadruplet codons can improve their decoding efficiency in genetic circuits, which allowed us to develop a system for the programmable biosynthesis of exotic macrocyclic peptides in cells.
Dedifferentiating myoblasts back into a satellite cell state in vitro
Satellite cells, the stem cells of skeletal muscle, are responsible for muscle development and regeneration. Although low in abundance, satellite cells can be isolated from muscle but cannot be propagated successfully in culture in numbers needed for therapeutic use. We developed a method to generate cells with satellite cell characteristics from skeletal muscle organoid cultures.
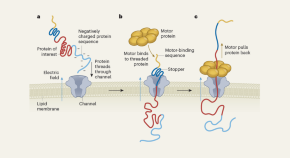
Thread, read, rewind, repeat: towards using nanopores for protein sequencing
Biologists would love to be able to read out the amino-acid sequence of any protein molecule as they would the letters of a sentence. A system in which a biological motor pulls proteins through a pore brings the dream closer to reality.
- Aleksei Aksimentiev
Related Subjects
- Animal biotechnology
- Applied immunology
- Assay systems
- Biomaterials
- Biomimetics
- Cell delivery
- Environmental biotechnology
- Expression systems
- Functional genomics
- Gene delivery
- Gene therapy
- Industrial microbiology
- Metabolic engineering
- Metabolomics
- Molecular engineering
- Nanobiotechnology
- Nucleic-acid therapeutics
- Oligo delivery
- Peptide delivery
- Plant biotechnology
- Protein delivery
- Regenerative medicine
- Stem-cell biotechnology
- Tissue engineering
Latest Research and Reviews
Facile photopatterning of perfusable microchannels in hydrogels for microphysiological systems
A cost-effective, facile, versatile and ultrafast methodology to fabricate perfusable microchannels of complex shapes in photopolymerizable hydrogels without the need for specialized equipment or sophisticated protocols.
- Ana Mora-Boza
- Adriana Mulero-Russe
- Andrés J. García
Engineering potent chimeric antigen receptor T cells by programming signaling during T-cell activation
- Aileen W. Li
- Jessica D. Briones
- Alexander S. Cheung
Plug-and-play protein biosensors using aptamer-regulated in vitro transcription
Biosensors that accurately detect proteins are critical for biological applications, but modular transduction of binding into useful outputs is a challenge. Here, authors develop a modular platform using aptamer-regulated transcription to detect proteins and process RNA outputs via molecular circuits.
- Heonjoon Lee
- Rebecca Schulman
Expanding the CRISPR base editing toolbox in Drosophila melanogaster
Testing three types of cytosine base editors in fruit flies demonstrates the potential usage of these base editors in genetic biocontrol strategies, with the rAPO-1 achieving high editing efficiency (~99%) with no detectable INDEL mutations.
- Michael Clark
- Christina Nguyen
- Víctor López Del Amo
Chemiosmotic nutrient transport in synthetic cells powered by electrogenic antiport coupled to decarboxylation
Replicating natural processes in synthetic cells is key to further development and understanding. Here, the authors develop a synthetic reaction network for the generation of metabolic energy in the form of proton motive force, used to drive the accumulation of nutrients and enable internal metabolism in cell-like vesicles.
- Miyer F. Patiño-Ruiz
- Zaid Ramdhan Anshari
- Bert Poolman
Tuning extracellular fluid viscosity to enhance transfection efficiency
Gene therapies and cellular programming rely on effective cell transfection. Here it is shown that optimizing the viscosity of cell culture media to match that of biological fluids substantially enhances the transfection efficiency for various gene delivery vehicles across different cell types.
- Hai-Quan Mao
News and Comment
Human intestinal immuno-organoids.
- Chiara Anania
Wobble base improves precision in RNA editing

I make fake eyes for those who need them
As the only ocularist in Uganda, Franklin Wasswa produces customized prosthetic eyes for people who’ve lost their own to injury or disease.
- Linda Nordling
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies

COMMENTS
Discover the latest research and news on plant biotechnology from Nature Portfolio, covering topics such as genetic engineering, tissue culture and biofuels.
Plant biotechnology is a mature technology. Modern biotechnology was a scientifically obvious outcome of the striking advances in molecular biology that followed the discovery of the bacterial DNA restriction-modification system (Luria and Human, 1952; Luria, 1953; Dussoix and Arber, 1962; Nathans and Smith, 1975).Microorganisms and plants were the first organisms to be manipulated to serve ...
Abstract. Biotechnology explores the metabolic properties of living organisms for the production of valuable products of a very different structural and organizational level. Plant serves as an important source of primary and secondary metabolites used in pharmacy, biotechnology, and food technology. Plant biotechnology has gained importance in ...
Plant Biotechnology Journal presents research at the forefront of applied plant science and molecular plant sciences. Published in partnership with the Society for Experimental Biology (SEB) and the Association of Applied Biology (AAB) it is dedicated to showcasing original research and insightful reviews by renowned researchers in the field of plant biotechnology.
Read the latest Research articles in Plant biotechnology from Nature Biotechnology. ... Plant biotechnology articles within Nature Biotechnology. Featured. Research Briefing | 20 May 2024.
a, Agrobacterium-mediated transformation strategies include callus induction followed by organogenesis (traditional transformation), somatic embryogenesis and de novo shoot regeneration in planta ...
Department of Plant and Microbial Biology, Program in Genetics, North Carolina State University, Raleigh, NC, United States. Plant biology is a key area of science that bears major weight in the mankind's ongoing and future efforts to combat the consequences of global warming, climate change, pollution, and population growth.
The Plant Biotechnology section at Frontiers in Plant Science mainly publishes applied studies examining how plants can be improved using modern genetic techniques (Lloyd and Kossmann, 2021).This Research Topic was designed to allow editors from the section to highlight some of their own plant biotechnological work.
The Plant Biotechnology section at Frontiers in Plant Science mainly publishes applied studies examining how plants can be improved using modern genetic techniques (Lloyd and Kossmann, 2021). This Research Topic was designed to allow editors from the section to highlight some of their own plant biotechnological work.
Hence, research should be focused on improving current delivery methods or developing novel ones to facilitate CRISPR/Cas9-based gene editing studies. Strict regulations on the sale and commercial growth of gene-edited crops have restricted more efforts in applying CRISPR/Cas9 technology in plant species. Therefore, a shift in public viewpoint ...
Plant Biotechnology Reports is a peer-reviewed journal emphasizing fundamental and applied research in plant biotechnology. Offers comprehensive coverage extending to molecular biology, genetics, biochemistry, and more. Prioritizes studies on plants indigenous to the Asia-Pacific region. Encourages studies related to commercialization of plant ...
Plant Biotechnology Journal is published by Wiley-Blackwell in association with the Society for Experimental Biology (SEB) and the Association of Applied Biology (AAB). Plant Biotechnology Journal aims to publish high-impact original research and incisive reviews by leading researchers in applied plant science, with an emphasis on molecular plant sciences and their applications through plant ...
The Plant Biotechnology section at Frontiers in Plant Science mainly publishes applied studies examining how plants can be improved using modern genetic techniques (Lloyd and Kossmann, 2021). This Research Topic was designed to allow editors from the section to highlight some of their own plant biotechnological work.
Kirakosyan's research on natural products of medicinal value in plants focuses on the molecular mechanism of secondary metabolite biosynthesis in selected medicinal plant models. His primary research interests focus on the uses of plant cell biotechnology to produce enhanced levels of medicinally important, value-added secondary metabolites ...
Here, we reviewed challenges in plant bioengineering and discussed opportunities in the emerging field of nano-based plant biotechnology to advance plant genetic engineering, as well as post ...
Plant biotechnology is a set of techniques used to adapt plants for specific needs or opportunities. Situations that combine multiple needs and opportunities are common. For example, a single crop may be required to provide sustainable food and healthful nutrition, protection of the environment, and opportunities for jobs and income. Finding or developing suitable plants is typically a highly ...
The future of plant biotechnology in a globalized and environmentally. endangered world. Marc Van Montagu 1. 1 VIB-International Plant Biotechnology Outreach, Ghent University, Ghent, Belgium ...
Wang's lab has been working with plant stem cells for years trying to understand how they control their stem cells, specifically the stem cells that give rise to vascular bundles - the structures that carry water and other nutrients throughout the plant. Recently the group published a paper in New Phytologist that sheds light on this question.
This section explores all branches of plant biotechnology, addressing the attempts of modern technologies to satisfy increasing demands for crop production. ... Submit your research. Start your submission and get more impact for your research by publishing with us. Author guidelines.
The purpose of this chapter is to provide a brief overview of the spectrum of applications of plant biotechnology that are in current use or are under development in research labs around the world. Plant biotechnology, in the sense of the application of recombinant DNA techniques to crop improvement, or the production of valuable molecules in ...
Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action. Fungal genome mining targeted to self-resistance genes close to biosynthetic gene clusters identifies a ...
Stem cell research is a hot topic. With applications for a host of human medical advancements, researchers have been working with animal and human stem cells for years. But animals aren't the only ...
In addition, it also offers a new insight of how these emerging methods will expedite advance research in plant biotechnology in the near future. Lastly, the various technological hurdles and inherent limitations of single-cell technology that need to be conquered to develop such outstanding possible knowledge gain is critically analyzed and ...
Here, we discuss the evolution of plant improvement, and how researchers leverage genomic analyses and revolutionary new plant breeding technologies like site-directed nucleases to enhance food ...
High-quality plant health research helps to protect cherished native species like the oak tree. Credit Paul Glendell, Natural England. My name's Alex and I am a scientist in the Plant Health Evidence and Analysis team. We use plant health research to fill any gaps in evidence and to inform Defra's policies.
Job Summary: The Wisconsin Crop Innovation Center (WCIC) at the University of Wisconsin-Madison is a ~100,000 square foot state-of-the-art agricultural biotechnology complex, housing laboratories and greenhouses dedicated to the production and analysis of transgenic and gene edited plants. This facility is the largest publicly held plant transformation facility in the United States and is ...
Biotechnology is a broad discipline in which biological processes, organisms, cells or cellular components are exploited to develop new technologies. New tools and products developed by ...